Fig. 5
- ID
- ZDB-FIG-201212-17
- Publication
- Issaka Salia et al., 2020 - Bioinformatic analysis and functional predictions of selected regeneration-associated transcripts expressed by zebrafish microglia
- Other Figures
- All Figure Page
- Back to All Figure Page
|
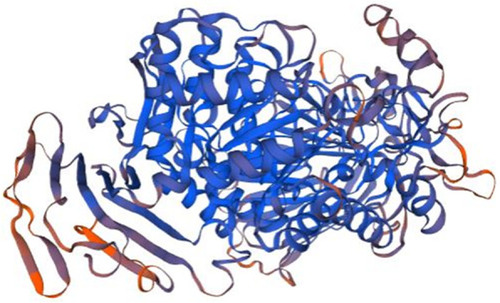
Homology model of P11. Maltase-glucoamylase, intestinal (3top.1.A in the rcsb protein database) is the template used for the homology modelling of P11. The X-RAY diffraction 2.9 Å was used to determine the experimental structure of 3top.1.A [ |