Figure 1
- ID
- ZDB-FIG-200910-1
- Publication
- Naert et al., 2020 - Maximizing CRISPR/Cas9 phenotype penetrance applying predictive modeling of editing outcomes in Xenopus and zebrafish embryos
- Other Figures
- All Figure Page
- Back to All Figure Page
|
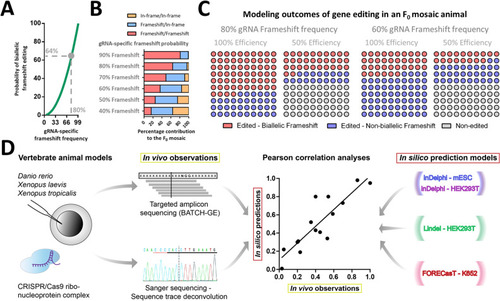
Theoretical models of how gRNA-specific efficiencies and frameshift gene editing outcome probabilities influence the cellular composition and percentage of protein knockout cells in a mosaic F0 animal model. ( |