Fig. 5
- ID
- ZDB-FIG-200323-17
- Publication
- Hoshijima et al., 2019 - Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
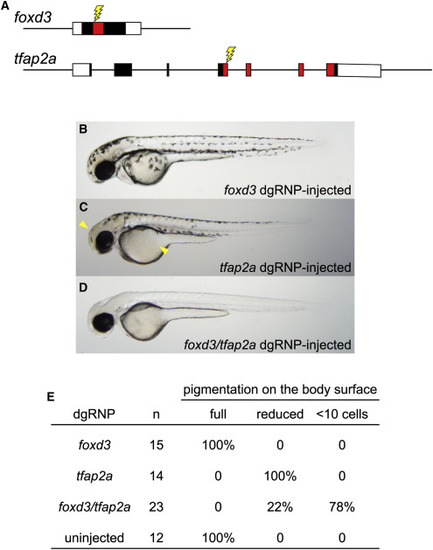
Redundant Gene Functions Can Be Uncovered by Targeting Two Genes Simultaneously with dgRNPs (A) dgRNPs were designed to target sequences (lightning bolts) encoding the DNA binding domains (red-shaded exon sequences) of the transcription factor products of the foxd3 and tfap2a genes. (B–E) Embryos targeted with either foxd3 dgRNPs or tfap2a dgRNPs resemble single mutants: (B) embryos targeted with foxd3 dgRNPs appear morphologically near-normal at 2 dpf, and (C) embryos targeted with tfap2a dgRNPs have moderately reduced pigmentation most notable on the head and yolk (arrowheads). (D) Most doubly injected embryos were devoid of melanophores at 2 dpf, resembling true foxd3;tfap2a double mutants (Arduini et al., 2009). (E) Three-fourths of the doubly injected embryos were devoid of pigment; the remaining embryos had markedly reduced pigmentation, consistent with incomplete loss of function of the targeted genes. |
Reprinted from Developmental Cell, 51(5), Hoshijima, K., Jurynec, M.J., Klatt Shaw, D., Jacobi, A.M., Behlke, M.A., Grunwald, D.J., Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish, 645-657.e4, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell