|
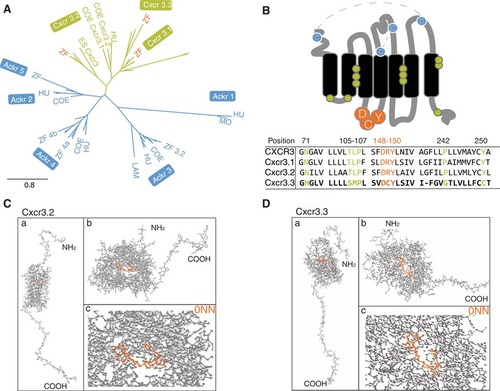
Cxcr3.3 has features of both conventional Cxcr3 receptors and ACKRs. Phylogenetic analyses including CXCR3 (green) and ACKR sequences (blue) of relevant species revealed that Cxcr3.3 is closely related to its paralogs Cxcr3.1 and Cxcr3.2 (A) (ZF, zebrafish; COE, coelacanth; HU, human; MO, mouse; ES, elephant shark; LAM, lamprey) despite having structural features of ACKRs (B), such as an altered E/DRY‐motif (orange) and microswitches (green). The predicted primary ligand‐binding site of both Cxcr3.2 and Cxcr3.3 is highly conserved and structural predictions suggest that they share several ligands (Supplementary Table 2). (C and D) The whole predicted structure of the Cxcr3.2 and Cxcr3.3 receptors (a), the ligand binding site of both proteins (b) and the binding of one of the shared predicted ligands (0NN) by each receptor (c)
|