Fig. 1
- ID
- ZDB-FIG-181207-15
- Publication
- Chong et al., 2018 - Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo
- Other Figures
- All Figure Page
- Back to All Figure Page
|
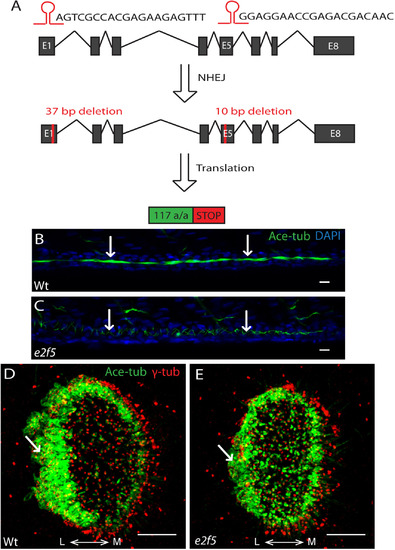
E2f5 is required for the formation of MCCs in the pronephric duct. A: Schematic showing CRISPR/Cas9-mediated genome editing at the zebrafish e2f5 locus using two guide RNAs targeting exon 1 and exon 5. After non-homologous end joining (NHEJ), F1 carriers contained a 37-bp deletion in exon 1, and a subsequent 10-bp deletion in exon 5. This translates to a truncated protein of 117-amino acid. B: Kidney tubule of a 48 hpf wild-type embryo populated with clusters of MCCs (arrows). C: Kidney tubule of an e2f5 mutant embryo completely devoid of MCCs. Cilia from monociliated cells, which are not affected by the loss of e2f5, are indicated (arrows). Acetylated-tubulin (Ace-tub; green), DAPI (blue). Scale bars= 10 µm. D: MCCs around the lateral rim of a nasal placode of a wild-type embryo at 72 hpf (arrow). E: Reduction in the number of nasal placode MCCs in e2f5 homozygous mutant (arrow). Acetylated-tubulin (Ace-tub; green); γ-tubulin (γ-tub; red). L: lateral; M: medial. Scale bars = 10 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article). |
| Antibody: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Protruding-mouth |
| Fish: | |
|---|---|
| Observed In: | |
| Stage Range: | Long-pec to Protruding-mouth |
Reprinted from Developmental Biology, 443(2), Chong, Y.L., Zhang, Y., Zhou, F., Roy, S., Distinct requirements of E2f4 versus E2f5 activity for multiciliated cell development in the zebrafish embryo, 165-172, Copyright (2018) with permission from Elsevier. Full text @ Dev. Biol.