Fig. 1
- ID
- ZDB-FIG-180709-2
- Publication
- Dobrzycki et al., 2018 - An optimised pipeline for parallel image-based quantification of gene expression and genotyping after in situ hybridisation.
- Other Figures
- All Figure Page
- Back to All Figure Page
|
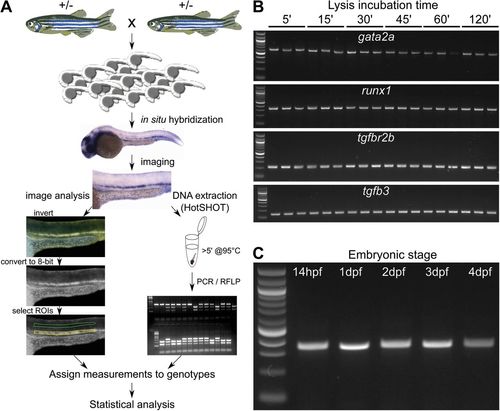
Overview of the method to extract DNA for genotyping zebrafish mutants after ISH and measure the mRNA levels. (A) Embryos collected from an incross of fish heterozygous for a mutant allele are probed for the measured gene with a standard ISH protocol. After imaging in 100% glycerol, genomic DNA is extracted using the HotSHOT protocol by adding the lysis buffer directly to the embryo in a 0.2 ml PCR tube, followed by a ≥5 min incubation at 95°C. This DNA is used for genotyping of the embryos by PCR and restriction fragment length polymorphism (RFLP). In parallel, the images for each embryo are inverted and converted to 8-bit greyscale. ROIs of identical shape and size containing the ISH signal (yellow) and background (green) are manually selected and measured. The measurements, assigned to corresponding genotypes, are statistically analysed. (B) DNA fragments amplified from single fixed and ISH-probed embryos after incubation in HotSHOT lysis buffer for 5, 15, 30, 45, 60 and 120 min followed by JumpStart™ REDTaq® ReadyMix™ PCR. Primer pairs designed for the gata2a, runx1, tgfbr2b and tgfb3 produced 600, 300, 176 and 144 bp fragments, respectively. Three wild-type embryos were used for each time point. First lane from the left: 100 bp DNA ladder. (C) DNA fragments amplified from single fixed and ISH-probed wild-type embryos after DNA extraction in HotSHOT lysis buffer for 5 min using a primer pair to amplify a 300 bp fragment of the runx1 locus with JumpStart™ REDTaq® ReadyMix™ PCR. First lane from the left: 100 bp DNA ladder. The lanes after the ladder represent embryos that are 14 h, 1, 2, 3 and 4 days old and have been stored in glycerol at room temperature for 2 years. |