Fig. 4
- ID
- ZDB-FIG-180413-12
- Publication
- De Kumar et al., 2017 - Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
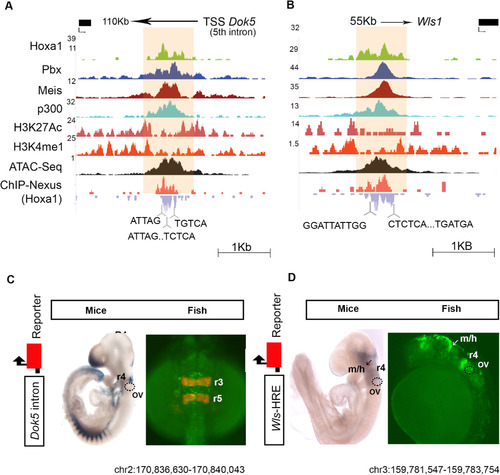
Hoxa1-bound regions function as enhancers. (A, B) Browser shots of two Hoxa1 bound regions (A, Dok5) and (B, Wls1) along with occupancy of TALE cofactors (Pbx and Meis), co-activator p300, modified histone marks characteristic of enhancers (H3K4me1 and H3K27Ac) and open chromatin states (ATAC-seq) are shown. ChIP-nexus data reveals the presence of multiple binding motifs (bottom) for Hoxa1 and its cofactors in the protected regions. (C, D) Regulatory analysis of the (A, Dok5) and (B, Wls1) Hoxa1-bound regions along with 250 bp flanking sequences in mouse and zebrafish embryos using LacZ and GFP transient transgenic reporter assays, respectively. In C, zebrafish embryos with mCherry inserted in the endogenous egr2b locus (krox20), mark r3 and r5 with reporter expression (Distel et al., 2009). The bound region from each gene mediates neural-specific expression in mouse and zebrafish embryos, indicating conservation of activity. OV, otic vesicle, m/h, mid/hindbrain boundary and r, >rhombomere. |
Reprinted from Developmental Biology, 432(1), De Kumar, B., Parker, H.J., Paulson, A., Parrish, M.E., Zeitlinger, J., Krumlauf, R., Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis, 151-164, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.