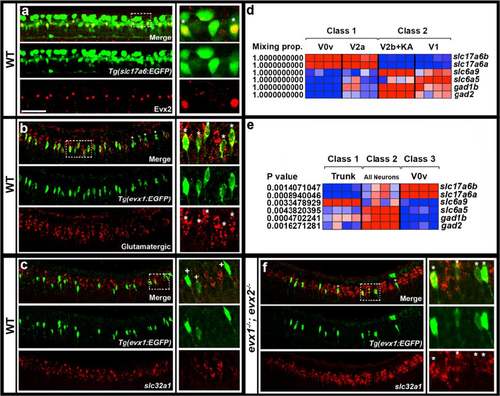
Neurotransmitter phenotypes of V0v cells. Lateral views of spinal cord at 27 h (a & b) or 30 h (c & f). All panels contain merged and single channel views. Smaller images on RHS are single confocal planes of white box regions. Double and single-labeled EGFP-positive cells are indicated in single confocal planes with stars and crosses respectively. a EGFP (green) and Evx2 (red) expression in Tg(slc17a6:EGFP) embryo. All Evx2-positive cells co-express EGFP. b EGFP (green) and glutamatergic marker (slc17a6b & slc17a6a; red) expression in Tg(evx1:EGFP) SU1 embryo. Occasional single-positive cells are indicated with crosses. These may be expressing glutamatergic markers at levels too low to detect (slc17a6 probes are weak in double-labeling experiments). Remaining cells are double-labeled. c & f EGFP (green) and slc32a1 (red) expression in Tg(evx1:EGFP) SU2 WT (c) and evx1;evx2 double mutant (f) embryos. No V0v cells are inhibitory in WT embryos, but most V0v cells are inhibitory in double mutants (occasional single-labeled cells are indicated with a cross in F). (d & e) Relative expression profiles of genes (names on right) indicative of neurotransmitter fates at 27 h. Columns represent individual microarray experiments. Rows indicate relative expression levels as normalized data transformed to mean of zero and standard deviation of +1 (highly-expressed, red) to -1 (weakly/not expressed, blue) sigma units. For details of how cells were isolated see methods. d Two-class eBayes comparison of excitatory (class 1) versus inhibitory (class 2) cells. Mixing proportion measures posterior probability, or likelihood that genes are differentially expressed (1 = highest probability of differential expression). e Three-class ANOVA comparison of V0v cells (class 3) versus trunk cells (class 1) and all post-mitotic neurons (class 2). P values test hypothesis that there is no differential expression between the 3 classes. V0v cells express glutamatergic markers slc17a6a and slc17a6b and do not express glycinergic or GABAergic markers in both comparisons. Scale bar: 50 μm (a-c & f)
|

