Fig. 1
- ID
- ZDB-FIG-171106-18
- Publication
- Son et al., 2016 - Transgenic FingRs for Live Mapping of Synaptic Dynamics in Genetically-Defined Neurons
- Other Figures
- All Figure Page
- Back to All Figure Page
|
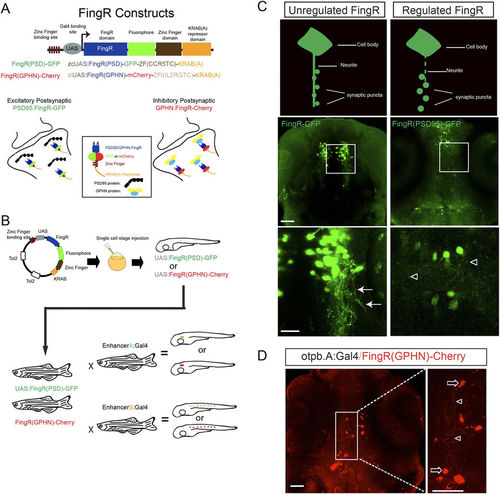
FingR constructs and testing in zebrafish. (A) Schematic diagram of FingR constructs used to generate plasmids and transgenic lines. FingR(PSD95)-GFP binds endogenous PSD-95 protein at the post-synaptic density; FingR(GPHN)-Cherry binds endogenous GPHN. (B) Use of inducible Gal4/UAS system with FingR(GPHN) or FingR(PSD95) in different Gal4 lines for differential synapse labeling (drawn by JHS). (C) Contrasting examples of unregulated (FingR-GFP) and regulated (FingR(PSD95)-GFP) FingR plasmids injected into Tg(otpb.A.Gal4) embryos. GFP signal from the unregulated FingR is distributed throughout the neuron and neurites (arrows) and individual synaptic puncta are not visualizable. In contrast, distinct puncta are seen (open arrowheads) using regulated FingR expression. Confocal images, scale bar 50 μm, 10 μm in inset panels; confocal z-stacks, ventral views, rostral to the top. D) Demonstration of puncta labeling with regulated FingR for GPHN. Confocal z-stacks, ventral view, rostral to top, of Tg(otpb.A:Gal4) embryo injected with FingR(GPHN)-Cherry. Arrows, neuron soma; arrowheads, puncta. Scale bar 50 μm, 10 μm in inset panel. |