Fig. 1
- ID
- ZDB-FIG-171011-12
- Publication
- Morrow et al., 2017 - tbx6l and tbx16 are redundantly required for posterior paraxial mesoderm formation during zebrafish embryogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
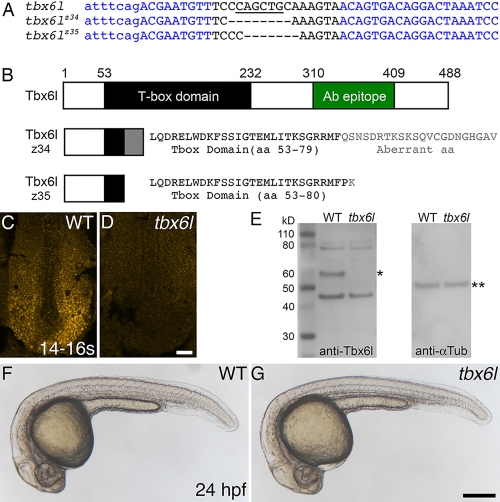
Zebrafish tbx6l mutants do not express Tbx6l protein and are homozygous viable. A: Sequence of tbx6l showing left and right TALEN binding sites (blue) and the spacer region (black) for wild-type, tbx6lz34, and tbx6lz35 alleles. Uppercase letters indicate exon 3 coding sequence; lowercase letters indicate intron sequence. Dashes indicate deleted nucleotides. A PvuII site (underlined) is deleted in both mutant alleles. B: Diagram of wild-type Tbx6l protein and predicted truncated Tbx6l proteins encoded by each mutant allele, showing the T-box domain (black box and black text), aberrant sequence caused by the introduced frameshift in mutant alleles (gray box and gray text), and the immunogen peptide (green box). C,D: Dorsal views of 14- to 16-somite stage wild-type (C) and tbx6lz34 mutant (D) embryos processed by immunofluorescence with Tbx6l antibody. E: Western blot of protein extracts prepared from wild-type (WT) and tbx6z35 homozygous mutant embryos, probed sequentially with anti-Tbx6l antibody (left blot), then with anti-alpha Tubulin antibody as a loading control (right blot). The inferred position of Tbx6l, which runs slightly higher than the predicted 55-kD mass, is marked by a single asterisk, and alpha-Tubulin (50 kD) by double asterisks. F,G: Live images of 24-hpf wild-type (F) and tbx6lz35 mutant (G) embryos. Scale bar in D (for C,D) = 25 μm. Scale bar in G (for F,G) = 250 μm. |
| Fish: | |
|---|---|
| Observed In: | |
| Stage Range: | 14-19 somites to Prim-5 |