Fig. 7
- ID
- ZDB-FIG-151203-5
- Publication
- Kagermeier-Schenk et al., 2011 - Waif1/5T4 Inhibits Wnt/β-Catenin Signaling and Activates Noncanonical Wnt Pathways by Modifying LRP6 Subcellular Localization
- Other Figures
- All Figure Page
- Back to All Figure Page
|
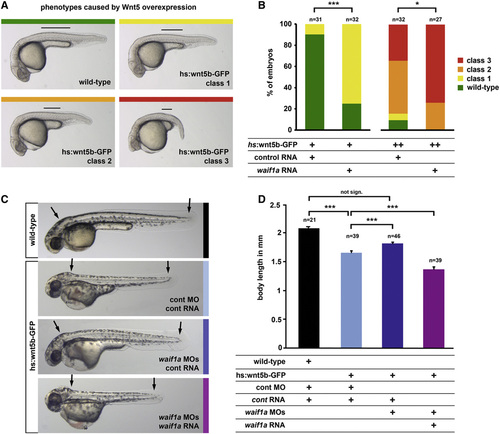
waif1a Enhances β-Catenin-Independent Wnt Signaling in Zebrafish Embryos (A) Classes of phenotypes induced in hsp70l:wnt5b-GFP transgenic zebrafish embryos heat shocked during gastrulation: class 1, shortening of the yolk extension (black line) by up to 1/3; class 2, up to 1/2; and class 3, more severe shortening plus other morphogenesis defects including cyclopia or open neural tube. (B) Phenotypic classes in waif1a RNA (75 pg) or RLuc (80 pg)-injected hsp70l:wnt5b-GFP embryos after weak induction (left, heat shock 39°C shield stage; p < 0.001, chi-square test) or strong induction (right, heat shock 40°C, 80% epiboly; p < 0.05, chi-square test). (C and D) hsp70l:wnt5-GFP embryos injected with standard control (cont) MO (8 ng) plus RLuc control RNA (30 pg), waif1a MO1 plus waif1a MO2 (5 ng each) plus RLuc RNA, or waif1a MOs plus MO-insensitive waif1a RNA (32 pg), heat shocked at 50% epiboly, and photographed at 48 hpf. Body length (from the otic vesicle to the tip of the tail, arrows) is plotted in (D). p < 0.001, Student′s t test. not sign., not significant. See also Figure S7. |
Reprinted from Developmental Cell, 21(6), Kagermeier-Schenk, B., Wehner, D., Ozhan-Kizil, G., Yamamoto, H., Li, J., Kirchner, K., Hoffmann, C., Stern, P., Kikuchi, A., Schambony, A., and Weidinger, G., Waif1/5T4 Inhibits Wnt/β-Catenin Signaling and Activates Noncanonical Wnt Pathways by Modifying LRP6 Subcellular Localization, 1129-43, Copyright (2011) with permission from Elsevier. Full text @ Dev. Cell