Fig. 1
- ID
- ZDB-FIG-130802-25
- Publication
- Harvey et al., 2013 - Identification of the zebrafish maternal and paternal transcriptomes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
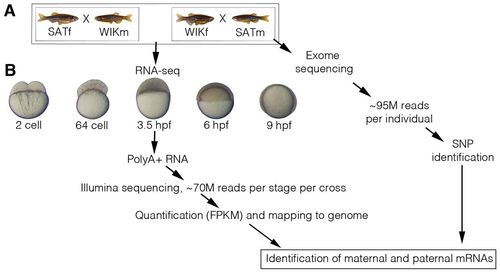
Identification of maternal and paternal mRNAs. (A) To identify maternal and paternal mRNAs, crosses of two different strains of zebrafish were used. A female SAT was crossed with a male WIK and the reciprocal cross of female WIK and male SAT was also performed. The four adults used were fin clipped and then subjected to exon enrichment and Illumina sequencing. On average, <95 million (M) reads were obtained for each sample/individual. Each sample was then run through our single-nucleotide polymorphism (SNP) calling pipeline. The exon-enriched sequences for the male and female in each cross were compared to identify homozygous SNPs that distinguished male and female alleles. (B) One hundred zebrafish embryos were collected at five different developmental stages from each cross: 2-cell, 64-cell, 3.5 hpf, 6 hpf and 9 hpf. Polyadenylated RNA was then extracted from total RNA and subjected to Illumina sequencing to produce <70 million reads per stage per cross. Reads were mapped to the zebrafish genome (Zv9) using TopHat and quantified using Cufflinks. SNPs identified were then used to identify maternal and paternal mRNAs. FPKM, fragments per kb per million reads; m, male; f, female. |