Fig. 5
- ID
- ZDB-FIG-110214-45
- Publication
- Ishioka et al., 2011 - Retinoic acid-dependent establishment of positional information in the hindbrain was conserved during vertebrate evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
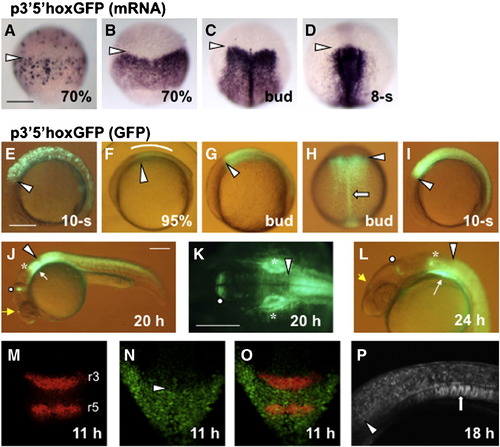
Early expression of hoxb1b in the posterior neural plate is driven by the downstream DNA. (A–D) Expression of 3′5′hoxGFP was visualized by WMISH using DIG-labeled probe in injected embryos (transient expression, A) or in Tg embryos (B–D). (E–P) Expression of 3′5′hoxGFP was detected as fluorescence under a stereomicroscope (E–L) or by confocal microscopy (M–P) in injected embryos (E) or in Tg embryos (F–P). (M-O) Expression of krox20 in r3 and r5 (M) was detected in a Tg embryo expressing 3′5′hoxGFP (N). Their merged image (O) shows that the anterior expression boundary of 3′5′hoxGFP coincides with the r3/r4 boundary. (A–D, H, K, M–O) Dorsal views with anterior to the top (A–D, H, M–O) or to the left (K). (E–G, I, J, L, P) Lateral views with anterior to the left. Anterior expression boundaries of 3′5′hoxGFP are shown with open arrowheads. The expression in the posterior hindbrain (F), pharyngeal arches, notochord, MHB otic vesicles, and diencephalon (J, L) are shown with a curve, white thin arrows, open thick arrows, open circles, asterisks, and yellow thin arrows, respectively. Scale, 200 μm. |
Reprinted from Developmental Biology, 350(1), Ishioka, A., Jindo, T., Kawanabe, T., Hatta, K., Parvin, M.S., Nikaido, M., Kuroyanagi, Y., Takeda, H., and Yamasu, K., Retinoic acid-dependent establishment of positional information in the hindbrain was conserved during vertebrate evolution, 154-168, Copyright (2011) with permission from Elsevier. Full text @ Dev. Biol.