Fig. 4
- ID
- ZDB-FIG-100302-14
- Publication
- Jing et al., 2010 - Temporal and spatial requirements of unplugged/MuSK function during zebrafish neuromuscular development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
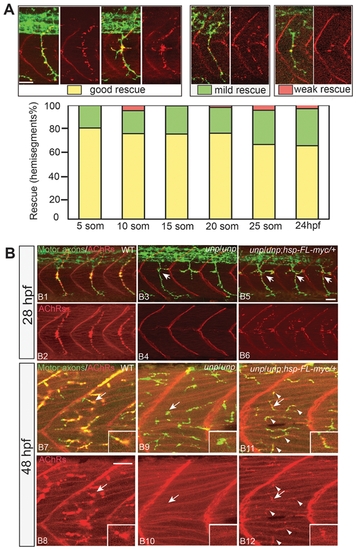
Rescue of neuromuscular synapses by unplugged/MuSK. (A) Rescue of neural synapses at different induction times by Tg(hsp70l:unplugged FL-myc). Embryos were heat-shocked starting at the indicated time points, fixed at 27hpf, and stained for motor axons (znp-1, green) and AChR clusters (α-BTX, red). Individual hemisegments were scored as ‘good rescue’ (when AChR clusters colocalized with presynaptic nerve ending on most muscle fibers, including along ectopic nerve ending), ‘mild rescue’ (when synapses formed on <50% of all muscles), or ‘weak rescue’ (when synapses formed only on non-migratory adaxial cells). 20 hemisegments were scored in each embryo. Results were summarized from two independent experiments (n = 160-380, average = 280, hemisegments per time point). (B) Tg(hsp70l:unplugged FL-myc) rescues neuromuscular synapses at 28 hpf and 48 hpf. Embryos were given a 40-min HS at 28hpf or 48hpf, and examined 30 minutes after the HS treatment. (B1–B6) In transgenic embryos, neuromuscular synapses formed on most muscle fibers and aligned with nerve ending. Arrows point to unplugged-characteristic pathfinding errors, indicating that the HS was too late to rescue axon pathfinding. (B7–B12) The single HS treatment at 48 hpf induced synapses in transgenic embryos (arrowheads in B11 and B12), albeit synapses appear smaller than those in wildtype. Inset in each panel is the enlarged image of the neuromuscular synapse pointed by the arrow. Scale bars: 20 μM. |