Fig. 2
- ID
- ZDB-FIG-081124-2
- Publication
- Madsen et al., 2008 - Zebrafish mutants calamity and catastrophe define critical pathways of gene-nutrient interactions in developmental copper metabolism
- Other Figures
- All Figure Page
- Back to All Figure Page
|
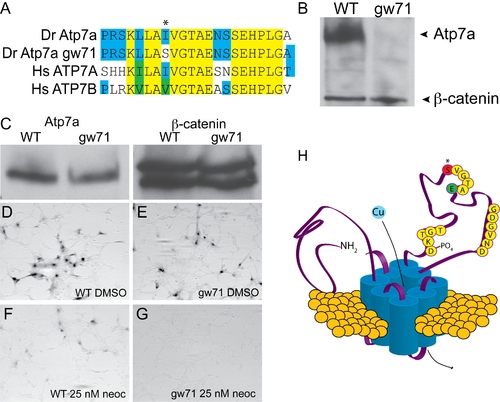
calamitygw71 contains a hypomorphic allele of atp7a. (A) The mutation in calgw71 is a T1061S substitution in a highly conserved region of the vertebrate copper transporters and exchanges a normally hydrophobic amino acid for a hydrophilic one (asterisk). (B) This single amino acid change results in near complete loss of immunoblot-detectable protein levels. Wild-type and gw71 mutant embryos were blotted for Atp7a using a peptide antibody to the C-terminus of the protein. β-catenin was used as a loading control. (C) Despite loss of protein in the zebrafish, transfection of an ATP7A-deficient human fibroblast cell line with either wild-type or mutant cDNA (derived from site-directed mutagensis of the wild-type) results in near equivalent expression of the zebrafish protein. β-catenin is again used as a loading control. (D–E) Both wild-type (D) and mutant (E) cDNAs are capable of producing functional protein as measured by functional tyrosinase activity in ATP7A deficient fibroblasts fixed and stained with L-DOPA. (F–G) Both wild-type (F) and mutant (G) Atp7a are sensitive to the effects of low-dose neocuproine in the above assay; however, the mutant cDNA is much more sensitive to mild copper chelation (F vs. G). (H) Model illustrating the relationship of the mutation (red, asterisk) to the known topology and functional domains of Atp7a. The mutation lies in the ATPase domain of the protein near a glutamate (green “E”) required for ATP binding and hydrolysis [10]. |
| Gene: | |
|---|---|
| Antibody: | |
| Fish: | |
| Anatomical Term: | |
| Stage: | Long-pec |