Fig. 1
- ID
- ZDB-FIG-080528-21
- Publication
- Lopez-Schier et al., 2005 - Supernumerary neuromasts in the posterior lateral line of zebrafish lacking peripheral glia
- Other Figures
- All Figure Page
- Back to All Figure Page
|
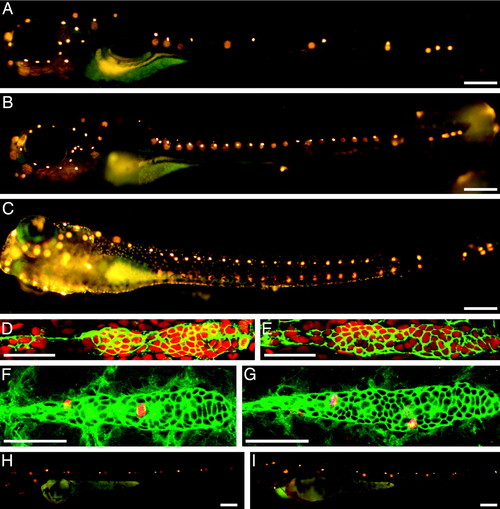
ngn1 mutant zebrafish develop extra neuromasts postembryonically. (A) When treated with the fluorescent compound 4-Di-2-ASP at 6 dpf, a wild-type larva displays only seven to nine neuromasts with functional hair cells on one side of its trunk. The neuromasts on the animal's opposite side appear as diffuse spots of fluorescence. (B) An ngn1 mutant at 6 dpf possesses more than twice as many neuromasts in its posterior lateral line. The number and pattern of neuromasts on the head is essentially normal, as is the animal's pigmentation. (C) At 6 dpf, an animal injected with a morpholino directed against the ngn1 transcript displays a pattern of supernumerary neuromasts comparable to that of an ngn1 mutant. Tilting of the larva separates the images of the neuromasts on the animal's two sides. (D) In a wild-type larva, an antibody against claudin b (green) labels the first primordium and the trail of cells left behind it; the nuclear stain TO-PRO 3 (red) marks all cells. (E) The primordium of an ngn1 mutant does not contain significantly more cells. (F) In a wild-type animal at 24 hpf, antiserum against α-tubulin strongly labels primordial cells (green), whereas that against phosphohistone H3 marks cells in mitosis (red). (G) The primordium is not more active mitotically in a homozygous ngn1 mutant larva. (H) When treated with 4-Di-2-ASP at 48 hpf, a wild-type animal shows only five or six neuromasts with functional hair cells on each side of its trunk. (I) At the same stage, an ngn1 mutant bears an equivalent number of mature neuromasts. In these and all subsequent illustrations, the animal's anterior is oriented to the left and its dorsum is situated to the top. (Scale bars, 50 μm for D–G and 100 µm for A–C, H, and I.) |