Fig. S1
- ID
- ZDB-FIG-071120-7
- Publication
- Peterkin et al., 2007 - Redundancy and evolution of GATA factor requirements in development of the myocardium
- Other Figures
- All Figure Page
- Back to All Figure Page
|
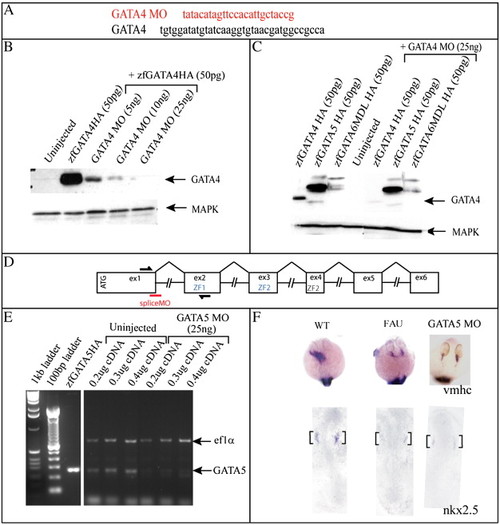
GATA4 MO specifically inhibits the translation of GATA4 RNA and GATA5 MO inhibits the production of endogenous GATA5 RNA. (A) GATA4 MO sequence and the ATG region to which it binds. (B) A Western blot to show the inhibition by GATA4 MO of translation of exogenous HA-tagged GATA4 RNA injected into Xenopus animal caps. (C) A Western blot to show that GATA4 MO specifically inhibits translation of GATA4 RNA injected into Xenopus oocytes, and has no effect on GATA5 or GATA6 translation. (D) Genomic structure of the GATA5 gene. Exons (ex) depicted as boxes, intron sizes not to scale. Splice MO binding site (red line) and primer binding sites for RT–PCR analysis (black lines) are shown. (E) The amplified 260 bp GATA5 fragment is present at a much lower level in the RNA extracted from embryos injected with 25 ng MO, indicating that there is a reduced level of correctly spliced GATA5 in the injected embryos. (F) Analysis of vmhc expression at 26 hpf (dorsal anterior view) and nkx2.5 expression at 10 somites (flat mount, dorsal view) shows that 25 ng GATA5 MO effectively phenocopues the fausttm236a mutant phenotype, black brackets identify cardiac expression. |
Reprinted from Developmental Biology, 311(2), Peterkin, T., Gibson, A., and Patient, R., Redundancy and evolution of GATA factor requirements in development of the myocardium, 623-635, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.