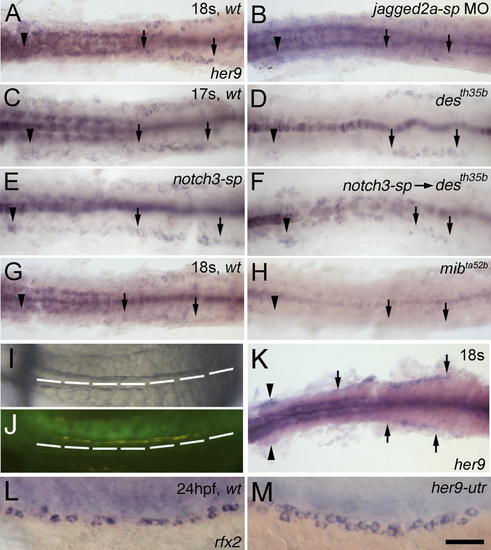
her9 is a Downstream Target Gene of Jagged2a-Notch1a/Notch3 Signaling(A and B) Compared to (A) wt embryos, her9 expression in the pronephric duct region at 18 ss is severely down-regulated in (B) jagged2a-sp morphants. (C–F) Compared to (C) wt embryos, her9 expression in the pronephric duct region at 17 ss is mildly down-regulated in (D) notch1a (desth35b) mutants and (E) notch3-sp morphants, and is severely down-regulated in (F) notch3-sp MO-injected notch1a (desth35b) mutants. (G and H) Compared to (G) wt embryos, her9 expression in the pronephric duct region at 18 ss is severely down-regulated in (H) mibta52b mutants. (I and J) Coinjection of GFP mRNA (50 pg) and notch1aicd mRNA (100 pg) into one blastomere at the two-cell stage leads to (I) somite boundary disruption in the right half of the embryo, while somites on the left side are segmented properly. (J) GFP expression demonstrates that mRNA is localized to the right half of the embryo. (K) Compared to the left side of the embryo, her9 expression in the duct (arrows) and glomerulus (arrowheads) is increased in the right side at 18 ss. (L and M) Compared to (L) wt embryos, the multi-cilia cell number is increased in (M) her9-utr morphants as shown by rfx2 expression at 24 hpf. All embryos, anterior to the left. (A–K) are dorsal views; (L and M) are lateral views. Bar scale: 100 μm (A–J), 130 μm (K), and 50 μm (L and M).
|

