Fig. 4
|
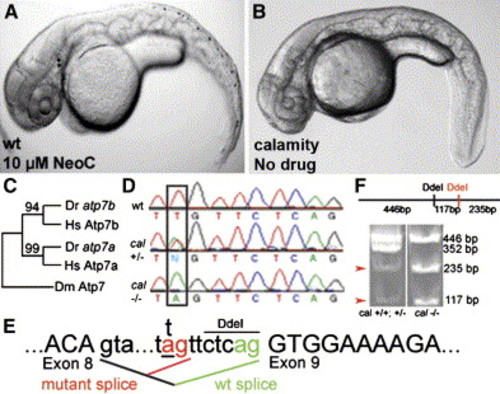
The zebrafish mutant calamityvu69 is defective in the zebrafish ortholog of the Menkes disease gene and is phenocopied by copper deficiency A and B) Copper-deficient embryo (A) and a cal homozygote (B) at 48 hpf. C and D) Phylogeny of copper-transport P type ATPases (C) displayed as maximum-likelihood tree with bootstrap values as shown. Aligned nucleotide sequence chromatograms (D) from genomic DNA from a portion of the atp7a gene from wild-type, cal heterozygote, and mutant embryos reveals a T→A transversion (box) in the intron 5′ to exon 9. E) Analysis of cDNA reverse transcribed from mRNA reveals that this mutation acts as a cryptic splice acceptor causing the insertion of seven nucleotides (TTCTCAG) into the mRNA. F) Restriction digests of a region amplified from atp7a cDNA reveal a novel DdeI site corresponding to the 7 bp insertion at the cal locus (arrowheads). |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Long-pec |
Reprinted from Cell Metabolism, 4(2), Mendelsohn, B.A., Yin, C., Johnson, S.L., Wilm, T.P., Solnica-Krezel, L., and Gitlin, J.D., Atp7a determines a hierarchy of copper metabolism essential for notochord development, 155-162, Copyright (2006) with permission from Elsevier. Full text @ Cell Metab.