- Title
-
Investigating the Role of Zebrafish Retinoschisin Homologs Rs1a and Rs1b During Retinal Development
- Authors
- van der Veen, I., Koster, C., Brink, J.B.T., Kamermans, M., Boon, C.J.F.
- Source
- Full text @ Dev. Neurobiol.
|
The alignments of the protein sequences of Rs1a ( |
|
|
|
|
|
|
|
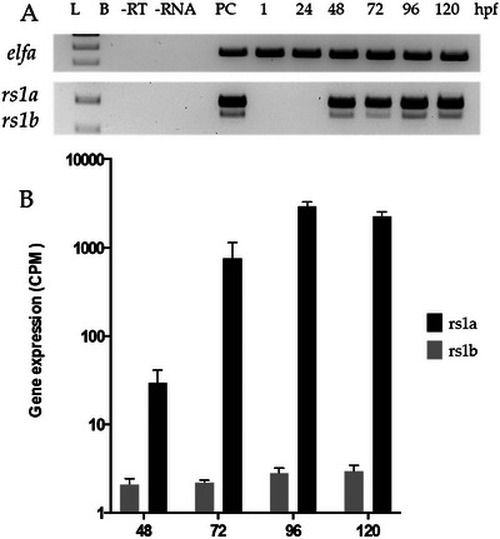
EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
|