- Title
-
A novel phex gene variant causes non-syndromic tooth agenesis
- Authors
- Pan, Y., Hua, B., Wang, H., Tan, S., Lu, T., Xiong, F., Ma, D.
- Source
- Full text @ BMC Oral Health
|
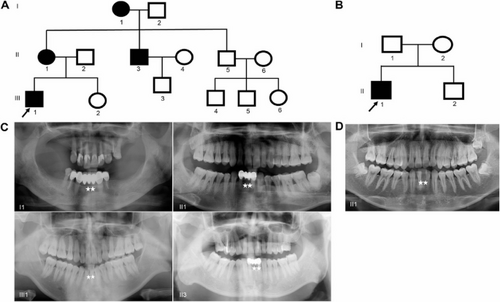
Pedigree and panoramic radiograph of family 1 (A) and family 2 (B). Males are marked as squares and females as circles. An arrow indicates the proband, and the black symbols indicate the affected individuals. (C-D) Panoramic radiograph of the patients shows all individuals had a congenital absence of mandibular central incisors. An asterisk marks the congenital missing tooth. Notably, Individual I1 has multiple missing teeth due to age, and the proband III1 has undergone orthodontic treatment |
|
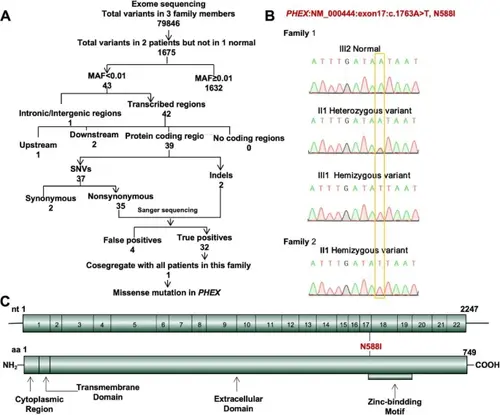
A-B Whole exome sequencing and Sanger sequencing revealed a novel PHEX variant (NM_000444, c.1763 A > T, N588I) identified in members with NSTA. C Location of the identified variant (marked in red) in schematic diagram of PHEX |
|
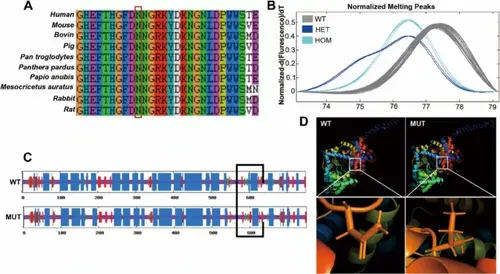
A Conservation analysis of the variation site among different species. B High-resolution melting (HRM) analysis of exon 17 of PHEX in 200 DNA specimens from random samples. C-D Protein prediction of the secondary and tertiary structure of PHEX |
|
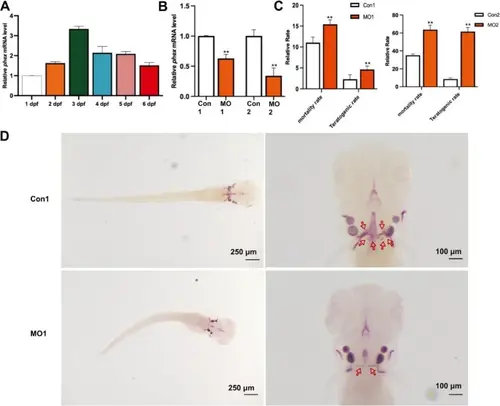
A Phex expression at different stages post-fertilization in zebrafish (n = 20 embryos per time point, 3 independent experiments). B Validation of Phex expression inhibition efficiency (n = 15 embryos per group, 3 independent experiments). C Statistics depicting mortality and malformation rates at varying concentrations of MO (n = 120 embryos total, pooled from 3 independent experiments with n = 40 per experiment). D Microinjection of zebrafish eggs and observation of tooth development at 6 days post-fertilization (dpf) via Alizarin Red staining (n = 30 embryos per group, 3 independent experiments). Scale bars: 250 μm (embryo images), 100 μm (tooth staining images). Data represent mean ± SEM. Statistical analysis performed using Student’s t-test for pairwise comparisons and one-way ANOVA for multiple group comparisons. *p < 0.05, **p < 0.01, ***p < 0.001 vs. control group PHENOTYPE:
|
|
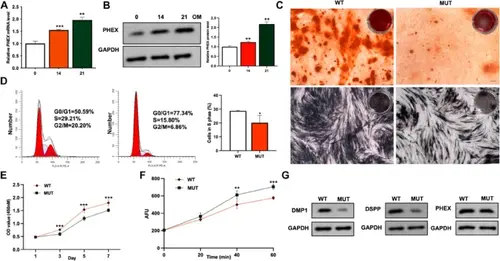
The qRT-PCR A and Western blot B results demonstrate upregulation of PHEX expression during mineralization induction in DPSCs (qRT-PCR: n = 3 per time point, 4 independent experiments; Western blot: n = 3 per time point, 3 independent experiments). C The results of ARS and ALP staining reveal reduced mineralization capability in mutant cells (ARS: n = 6 per group, 4 independent experiments; ALP: n = 6 per group, 3 independent experiments). Cell cycle analysis D and CCK-8 assay E demonstrate reduced cell proliferation capability after mutation (Cell cycle: n = 3 per group, 3 independent experiments; CCK-8: n = 6 per group, 4 independent experiments). F PHEX enzyme activity assay showed enhanced enzymatic activity in the MUT group (n = 4 per group, 3 independent experiments). G Western blot results demonstrate that after mutation, the expression of PHEX enzyme substrates DMP1 and OPN is downregulated, while the expression of PHEX itself remains unchanged (n = 3 per group, 3 independent experiments). Data represent mean ± SEM. Statistical comparisons performed using Student’s t-test for two-group comparisons and one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001 vs. control group (DPSCs-WT) |
|
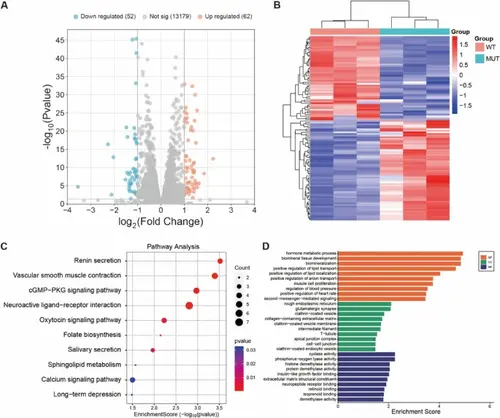
A Volcano plot of differentially expressed genes (n = 3 technical replicates per group, single RNA-seq experiment). Green dots represent down-regulated genes, and red dots represent up-regulated genes (|log2FC|≥ 1 and adjusted p ≤ 0.05). B Heatmap showing the expression levels of transcripts in samples from the DPSCs-MUT and DPSCs-WT (n = 3 technical replicates per group). Red color refers to upregulation, and blue color refers to downregulation of gene transcription. C KEGG pathway analysis ranked the top 10 KEGG pathways (|log2FC|≥ 1 and adjusted p ≤ 0.05, based on 3 technical replicates per group). D Go results of biological process, cellular component, and molecular function (enrichment analysis based on differentially expressed genes with adjusted p ≤ 0.05, using 3 technical replicates per group). RNA sequencing was performed using Illumina platform with > 20 million clean reads per sample. Differential expression analysis was conducted using DESeq2 with Benjamini-Hochberg correction for multiple testing. Statistical significance: *adjusted p < 0.05, **adjusted p < 0.01, ***adjusted p < 0.001 |