- Title
-
A conserved domain of Cfap298 governs left-right symmetry breaking in vertebrates
- Authors
- Cortez, M., Young, C.B., Little, K.A., Grimes, D.T., Devenport, D., Burdine, R.D.
- Source
- Full text @ J. Cell Sci.
|
Cfap298ΔΔS mutants display left–right laterality defects. (A) Generation of the Cfap298ΔΔS mutant allele by targeting exon 4 of the Cfap298 gene using CRISPR/Cas9 gene targeting. Guide recognition sequence within the exon 4 sequence is in red on the wild-type sequence. The Cfap298ΔΔS mutation consists of a 6 base deletion and a base substitution of T→C, resulting in the removal of two amino acids at positions 161 an 162, and a missense mutation of P163S (gray shading). (B) AlphaFold predicted structures of CFAP298 and CFAP298ΔΔS proteins in the region of the ΔΔS mutation. Y161 and D162 are labeled in wild-type CFAP298, and S161 is labeled on CFAP298ΔΔS (Abramson et al., 2024). (C) Whole-embryo images of E14.5 wild-type and Cfap298ΔΔS/ΔΔS embryos. Scale bar: 1 mm. (D) Viability distribution of wild-type, heterozygous and homozygous mutant embryos at E9.5 (n=17), E13.5 (n=15), E14.5 (n=33), E15.5 (n=21) and E16.5 (n=6). (E) Representative images of body cavities showing heart, lungs and stomach positions from Cfap298ΔΔS/+ and Cfap298ΔΔS/ΔΔS E14.5 embryos. Scale bars: 1 mm. (F) Quantification of situs solitus, situs inversus and heterotaxy from wild-type (n=21), Cfap298ΔΔS/+ (n=46) and Cfap298ΔΔS/ΔΔS (n=18) embryos from E13.5–E15.5 stages. (G) Representative images of heart, lung and stomach from E14.5 Cfap298ΔΔS/+ and Cfap298ΔΔS/ΔΔS embryos displaying with situs solitus (normal), situs inversus (reversed) and heterotaxy phenotypes. Hearts and stomachs are outlined. Individual lung lobes are outlined and labeled with their position on either left (L) or right (R). Atria (red asterisks) and ventricle (black asterisk) are indicated in the heart with heterotaxy. Scale bars: 1 mm. |
|
Cfap298ΔΔS mutants have perturbed left–right patterning. (A–B‴) Representative HCR images for Nodal (green), Pitx2 (magenta) and Lefty1 (yellow) expression in a whole-mount E8.5 wild-type embryo (A) and Cfap298ΔΔS mutants (B′–B‴). Wild-type embryos exhibit left-sided Nodal and Pitx2 expression in the lateral plate mesoderm (LPM) (arrowheads) with Lefty1 expression along the embryonic midline (asterisk). (B–B‴) Images from Cfap298ΔΔS mutants showing one example of right-sided Nodal, Lefty and Pitx2 expression along the LPM (arrowheads) (B′), an example where Nodal and Pitx2 are absent in the LPM and Lefty1 is expressed weakly at the midline (B″, asterisk), and an example of abnormal expression with reduced and bilateral Nodal, right-sided Pitx2 (arrowheads) and absent Lefty1 in the midline (asterisk) (B‴). Scale bars: 100 μm. (C) Diagram of an E8.5 wild-type embryo viewed from the dorsal side, anterior is up. Expanded view of a cross-section through the node is shown on the right. Representative images of E8.5 node labeled by HCR for Nodal (green), Lefty1 (yellow) and Cfap298 (magenta) transcripts. Cross-section (1) and planar (2 and 3) views of the node showing Nodal expression in the crown cells but not at the pit cells. Lefty1-expressing cells mark the floorplate of the neural tube are visible at the base of the pit of the node. Cfap298-expressing cells are visible throughout the node. Scale bars: 10 μm. |
|
Cfap298ΔΔS mutants develop normal nodes. (A) Representative immunofluorescent images of E8.5 nodes from wild-type and Cfap298ΔΔS mutant embryos. Embryos are stained for cilia with acetylated tubulin (green) and γ-tubulin (magenta) antibodies. For wild-type and mutant nodes, one plane is shown for the crown cells and another plane is shown for pit cells along with a z-projection of the whole node. (B) Images of cilia are shown for wild-type and mutant nodes. Quantifications of cilium length for wild-type (n=3 nodes, 150 cilia total) and mutant (n=3 nodes, 150 cilia total) embryos showing no significant difference (P=0.76 by unpaired t-test). (C) Images showing CELSR1 (cyan) and γ-tubulin (red) marking basal bodies in wild-type (+/+) and Cfap298ΔΔS mutant embryos. Boxes in i and ii indicate regions shown in images iii and iv, respectively. (D) Circular histograms display magnitude and orientation of CELSR1 polarity along the AP axis of wild-type (n=3 nodes, 99 cells total) and mutant (n=3, 218 cells total) embryos. Average polarity magnitudes from wild-type and mutant nodes were determined not to be significantly different (P=0.0691 by unpaired t-test). (E) Quantification of basal body polarity along the AP axis of the wild-type (n=3, 86 cells total) and mutant (n=3, 97 cells total) embryos showing no significant difference (P=0.2877 by Chi-squared test). ns, not significant (P>0.05). Scale bars: 10 μm. |
|
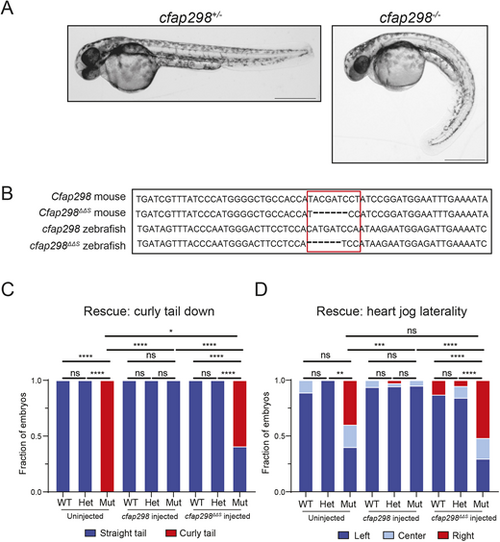
Cfap298ΔΔS does not rescue LR defects in zebrafish. (A) Representative images of cfap298+/− and cfap298−/− zebrafish embryos at 48 hpf, with cfap298+/− embryos displaying straight bodies and/or tails compared to cfap298−/−, which display curved bodies and/or tails. (B) Comparison of mouse wild-type Cfap298 and Cfap298ΔΔS with wild-type zebrafish cfap298 and engineered cfap298ΔΔS sequence. Area outlined in red indicates location of the cfap298ΔΔS mutation in mouse and zebrafish. (C,D) Quantification of body curvature (C) and heart position (D) observed in uninjected and injected embryos. Uninjected wild-type (n=9) and cfap298+/− (n=13) embryos were observed to have straight tails compared to cfap298−/− (n=10), which displayed statistically significant greater tail curvature (for wild type and cfap298+/−, P≥0.9999; for wild type and cfap298−/−, P≤0.0001; for cfap298+/− and cfap298−/−, P≤0.0001 by Fisher's exact test). However, for left–right defects, we did not observe a statistically significant difference between wild type, cfap298+/− or cfap298−/−, but we did observe a statistically significant difference between cfap298+/− and cfap298−/− (for wild type and cfap298+/−, P=0.4091; for wild type and cfap298−/−, P=0.0705; for cfap298+/− and cfap298−/−, P=0.0021 by Fisher's exact test). For embryos injected with 500 pg of wild-type cfap298 mRNA, we observed no significant difference between wild-type (n=16), cfap298+/−(n=36) or cfap298−/− (n=20) embryos (for wild type and cfap298+/−, P≥0.9999; for wild type and cfap298−/−, P≥0.9999; for cfap298+/− and cfap298−/−, P≥0.9999 by Fisher's exact test) as well as no significant difference in left–right defects (for wild type and cfap298+/−, P=0.6769; for wild type and cfap298−/−, P≥0.9999; for cfap298+/− and cfap298−/−, P≥0.9999 by Fisher's exact test). For embryos injected with 500 pg of cfap298ΔΔS mRNA, wild-type (n=23) and cfap298+/−(n=38) embryos were observed to have mostly straight tails compared to cfap298−/− (n=27), which displayed statistically significant greater tail curvature (for wild type and cfap298+/−, P≥0.9999; for wild type and cfap298−/−, P≤0.0001; for cfap298+/− and cfap298−/−, P≤0.0001 by Fisher's exact test) as well as significantly higher incidence in left–right defects (for wild type and cfap298+/−, P=0.2178; for wild type and cfap298−/−, P≤0.0001; for cfap298+/− and cfap298−/−, P≤0.0001 by Fisher's exact test). We also observed a significant difference in tail curvature between cfap298−/− uninjected, wild-type mRNA-injected and ΔΔS mRNA-injected embryos (for uninjected and wild-type mRNA injected, P≤0.0001; for uninjected and ΔΔS mRNA injected, P=0.0179; for wild-type mRNA injected and ΔΔS mRNA injected, P≤0.0001 by Fisher's exact test). We did not observe a significant difference in cfap298−/− uninjected and ΔΔS mRNA-injected embryos (P=0.8864 by Fisher's exact test), but we did observe a significant difference in left–right defects in cfap298−/− embryos injected with wild-type mRNA compared to uninjected embryos or embryos injected with ΔΔS mRNA (for uninjected and wild-type mRNA injected, P=0.0009; for wild-type mRNA injected and ΔΔS mRNA injected, P≤0.0001 by Fisher's exact test). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Scale bars: 500 μm. PHENOTYPE:
|