- Title
-
Second generation lethality in RNAseH2a knockout zebrafish
- Authors
- Thomas, R.C., Zaksauskaite, R., Al-Kandari, N.Y., Hyde, A.C., Abugable, A.A., El-Khamisy, S.F., van Eeden, F.J.
- Source
- Full text @ Nucleic Acids Res.
|
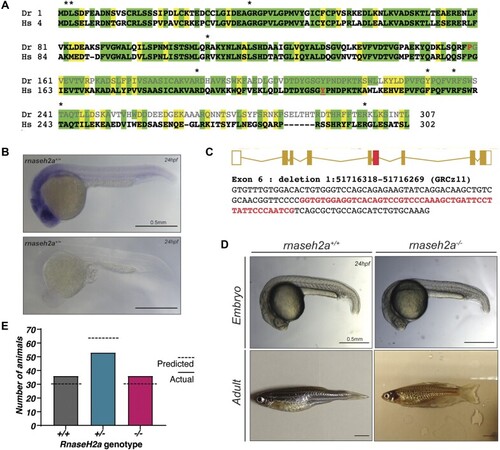
Generation of |
|
Offspring of |
|
|
|
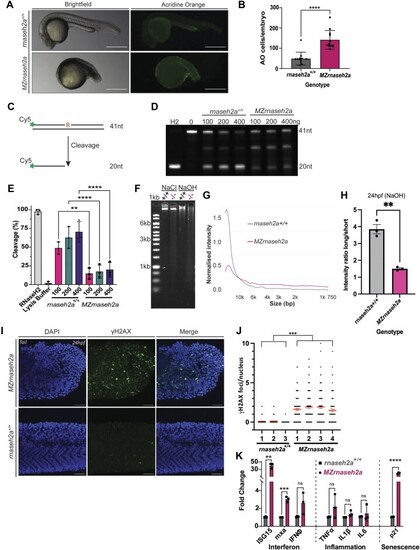
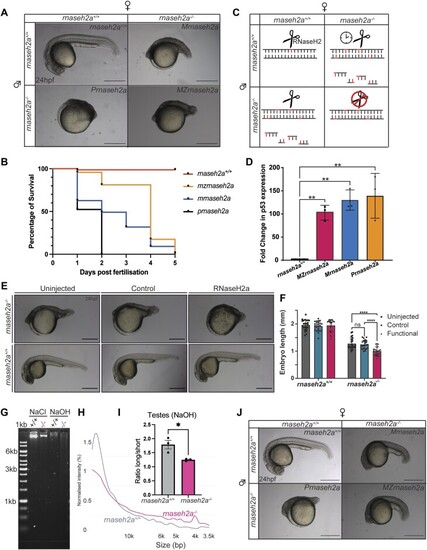
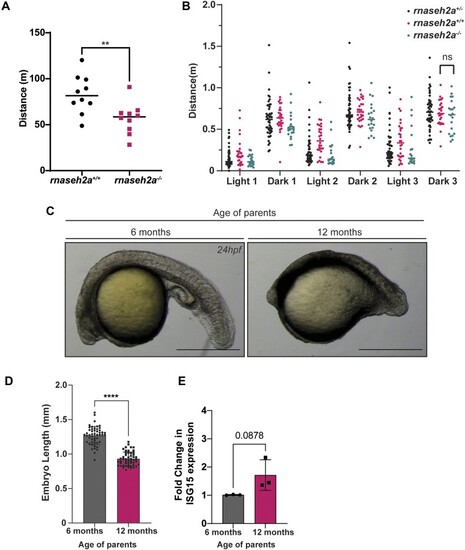
Re-introduction of RNaseH2a is unable to rescue the developmental phenotype. ( |
|
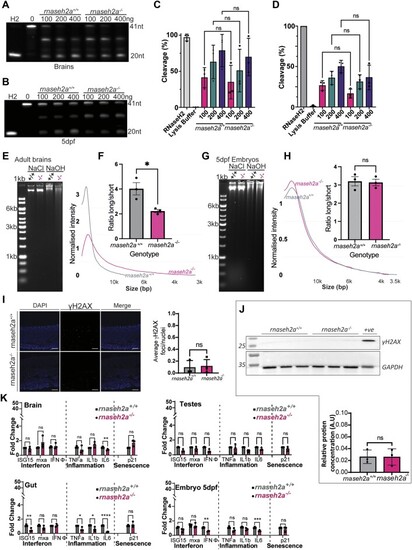
Embryos from older rnaseh2a−/− fish are significantly underdeveloped compared with embryos from younger fish. ( |