- Title
-
Starvation-resistant cavefish reveal conserved mechanisms of starvation-induced hepatic lipotoxicity
- Authors
- Pozo-Morales, M., Cobham, A.E., Centola, C., McKinney, M.C., Liu, P., Perazzolo, C., Lefort, A., Libert, F., Bai, H., Rohner, N., Singh, S.P.
- Source
- Full text @ Life Sci Alliance
|
Response of cavefish and surface fish larvae to starvation. |
|
Reduction in the size of liver upon accumulation of lipid droplets during starvation. |
|
|
|
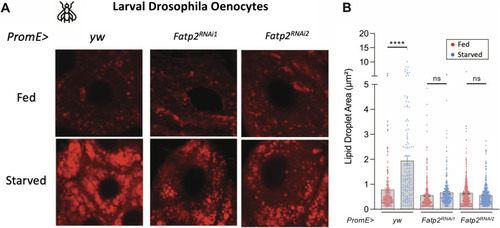
FATP2 is an evolutionarily conserved regulator of starvation-induced lipidosis. |
|
A representative chromatogram of the |
|
Macrophages in the liver of 8 dpf zebrafish upon normal feeding. |
|
Accumulation of hepatic lipid droplets after removal of lipofermata. |
|
Transcriptional changes in the liver of 6 dpf fasting zebrafish upon lipofermata treatment. |
|
Lipofermata treatment does not impact animal growth, liver size in fed animals, or starvation resistance. |