- Title
-
Angiopoietin 1 and integrin beta 1b are vital for zebrafish brain development
- Authors
- Chen, Y.C., Martins, T.A., Marchica, V., Panula, P.
- Source
- Full text @ Front. Cell. Neurosci.
|
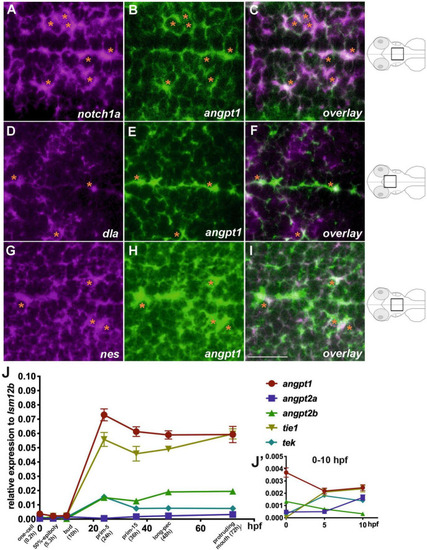
Co-localization of angpt1 with neurogenic markers in midline proliferative zone (shown schematically on the right). Double FISH in 2-dpf brain on 1.0 um thick optical sections using antisense RNA probes simultaneously hybridized against EXPRESSION / LABELING:
|
|
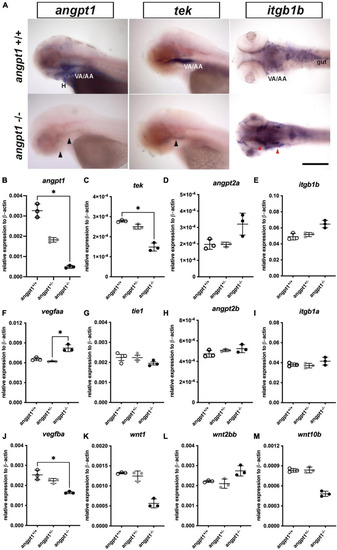
mRNA expression levels of angiogenic factors in 3-dpf EXPRESSION / LABELING:
PHENOTYPE:
|
|
mRNA expression levels of neurogenesis markers in 3-dpf EXPRESSION / LABELING:
PHENOTYPE:
|
|
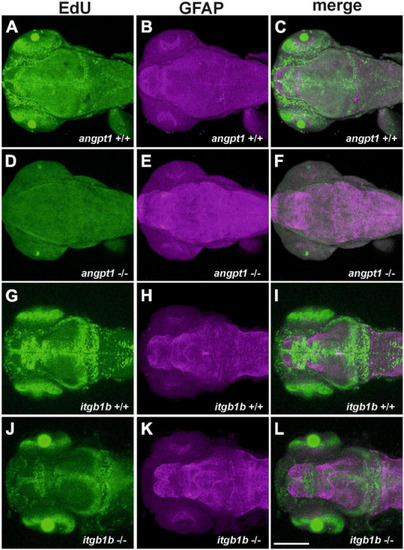
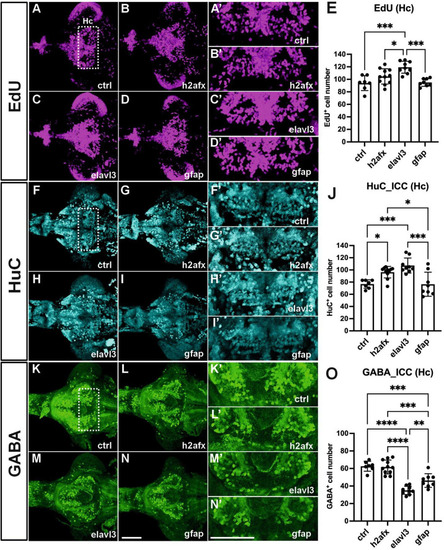
Decreased proliferation and increased gfap intensity in EXPRESSION / LABELING:
PHENOTYPE:
|
|
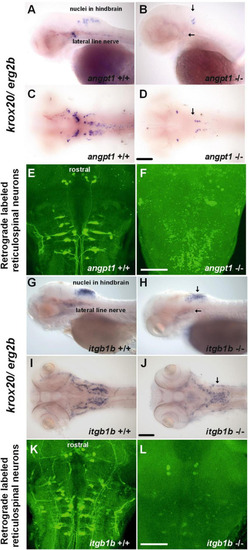
Deficiencies of reticulospinal neurons in PHENOTYPE:
|
|
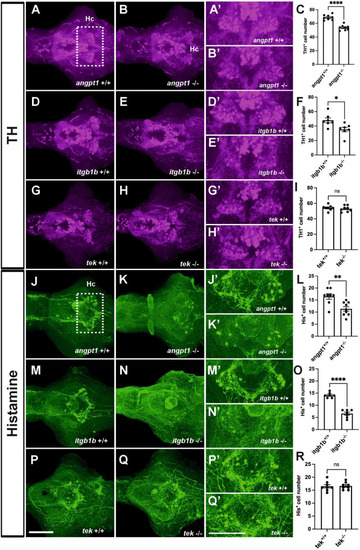
Deficient dopaminergic and histaminergic neurons are found in |
|
Transgenic expression of zebrafish |
|
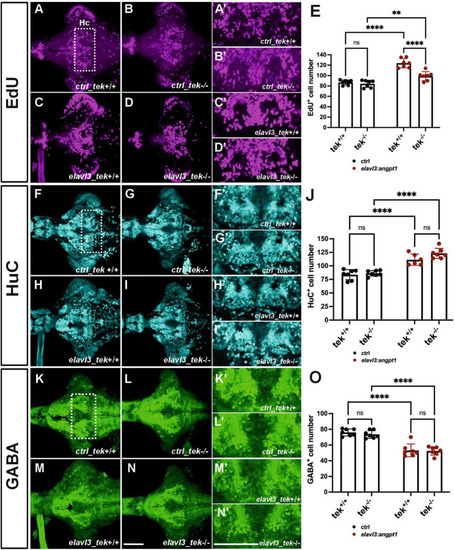
Zebrafish angpt1 regulates neurogenesis in a tek-independent manner. 5-dpf dissected brains were collected from |