- Title
-
Anti-diarrheal drug loperamide induces dysbiosis in zebrafish microbiota via bacterial inhibition
- Authors
- Stevick, R.J., Audrain, B., Bedu, S., Dray, N., Ghigo, J.M., Pérez-Pascual, D.
- Source
- Full text @ Microbiome
|
Experimental scheme of the larval zebrafish assays and sample collection. Conventional, mono-colonized, or mix5-colonized larval zebrafish were exposed at 5 dpf to water (control), DMSO (control) or 10 mg/L loperamide hydrochloride (treated) for 24 h and then transferred to water at 6 dpf. Samples were collected at 6 dpf, 7 dpf, and 11 dpf (T0, T1, T5) to measure fish growth and quantify bacterial community composition in all conditions ( |
|
Loperamide affects conventional zebrafish microbiota as measured by 16S rRNA gene amplicons. |
|
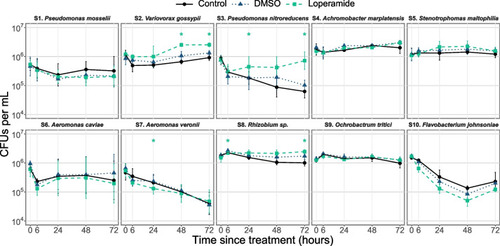
In vitro survival in water of zebrafish-associated bacterial strains is affected by loperamide. Survival in water for 72 h after inoculation at 106 CFUs/mL (mean ± standard deviation per condition is shown, |
|
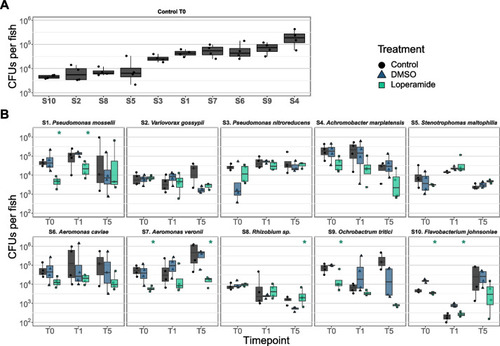
Loperamide can increase or reduce zebrafish mono-colonization. CFUs per fish of mono-colonized fish in |
|
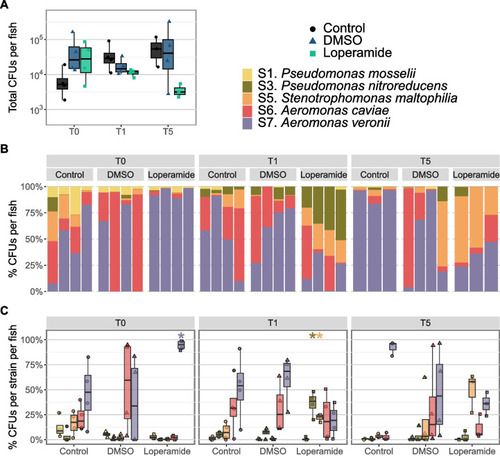
Loperamide affects mix5-colonized gnotobiotic zebrafish bacterial load and composition. |
|
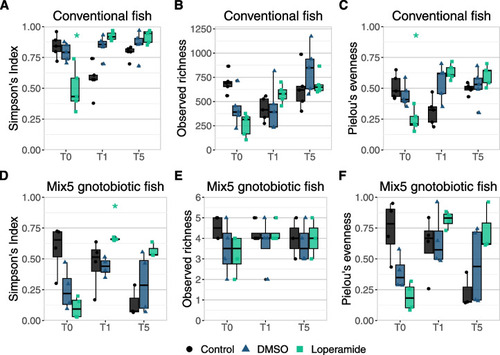
Simpson’s diversity of conventional and gnotobiotic zebrafish decreases but recovers after loperamide treatment. Alpha-diversity indices calculated at each timepoint for control water, DMSO, and loperamide-treated samples for |