- Title
-
Novel bi-allelic variants of CHMP1A contribute to pontocerebellar hypoplasia type 8: additional clinical and genetic evidence
- Authors
- He, T., Sun, H., Xu, B., Qu, H., Cai, X., Zhou, H., Liu, Y., Lin, Z., Zhang, X.
- Source
- Full text @ Front Neurol

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |
|
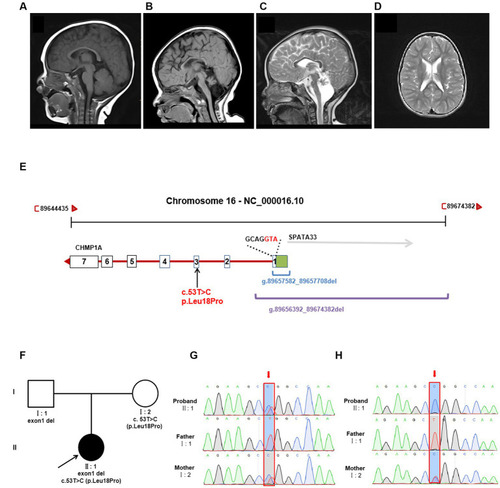
Brain MRI and genetic analysis for two novel |
|
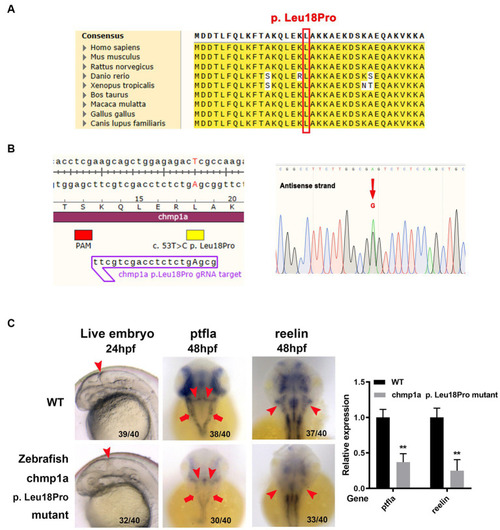
Zebrafish |