- Title
-
Generation and characterization of a zebrafish knockout model of abcb4, a homolog of the human multidrug efflux transporter P-glycoprotein
- Authors
- Park, J., Kim, H., Alabdalla, L., Mishra, S., Mchaourab, H.
- Source
- Full text @ Hum. Genomics
|
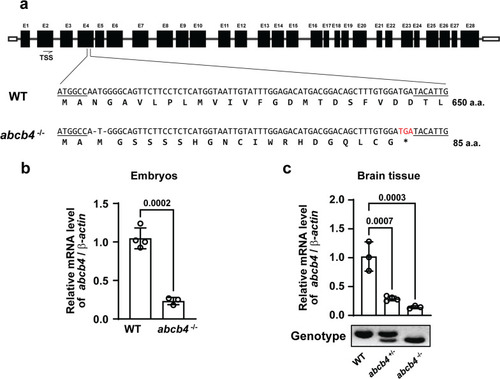
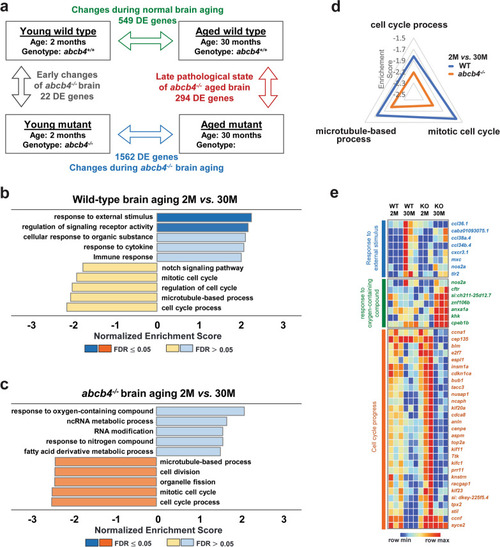
Generation of zebrafish |
|
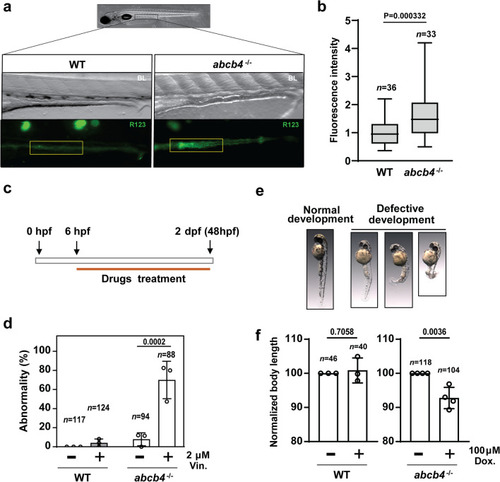
Zebrafish Abcb4 is an efflux transporter of rhodamine123, a substrate for human Pgp. |
|
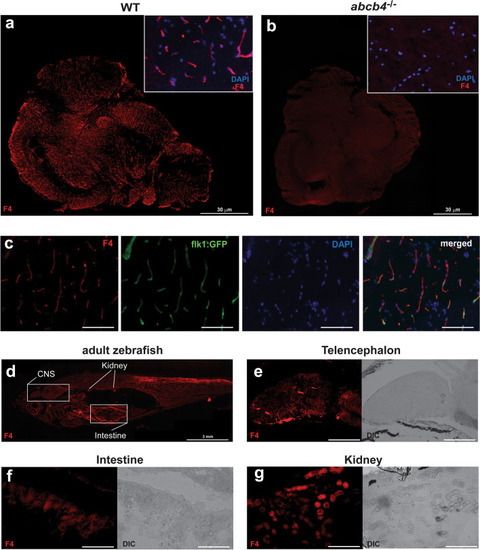
Zebrafish Abcb4 protein localizes to blood vessels in the zebrafish brain. Brain tissues of WT |
|
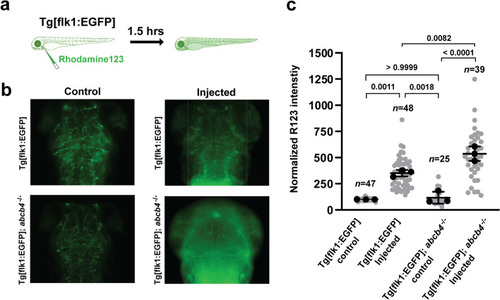
Efflux activity of zebrafish Abcb4 in the larval brain vasculature EXPRESSION / LABELING:
PHENOTYPE:
|
|
Age-related transcriptome profiling in |