- Title
-
Essential role of an ERV-derived Env38 protein in adaptive humoral immunity against an exogenous SVCV infection in a zebrafish model
- Authors
- Hong, Y., Hu, C.B., Bai, J., Fan, D.D., Lin, A.F., Xiang, L.X., Shao, J.Z.
- Source
- Full text @ PLoS Pathog.
|
Bioinformatic characterization of ERV-E5.1.38-DanRer element and Env38 molecule.
(A) Genomic localization of ERV-E5.1.38-DanRer element in zebrafish. The locus of ERV-E5.1.38-DanRer element on zebrafish chromosome 5 is indicated in red arrow. The ERV-E5.1.38-DanRer is distributed inside si:zfos-375h5.1. The upstream hspb1, ywhag1, tspoap1, supt4h1, hsf5 and downstream rpain, c1qbp, znhit3, myo19 genes of ERV-E5.1.38-DanRer are presented, and att means attachment site. (B) Schematic diagram showing the transcription and splicing of env38 gene, as well as the structural characterization of Env38 protein, in which a signal peptide (1–18 amino acid), a furin cleavage site (R/K-X-R/K-R, 227–230 amino acid), an immunosuppressive domain (ISD, 309–325 amino acid), a transmembrane (TM) domain (383–405 amino acid) and conserved C-X-X-C and C-X5/6/7-C motifs are presented. The LTR consists of U3, R, and U5. The transcription of env38 begins at 5’-LTR-R and ends at 3’-LTR-R. (C) The five predicted N-glycosylation sites in SU subunit and two in TM subunit are marked with ASN residues. SU: surface subunit; TM: transmembrane subunit. (D) Hydrophobicity analysis of Env38 protein showed that the fusion peptide next to the furin-cleavage site was partial hydrophobic. FP: fusion peptide; NHR: N-terminal heptad repeats; CHR: C-terminal heptad repeats; TM: transmembrane domain. (E) Tertiary structure of Env38 monomer modeled by I-TASSER. Prominent central α-helical coiled-coil (CC) structures were found in the TM subunit of Env38 monomer protein. The top five threading templates were 6vkm, 6rx3A, 6vkm, 6fgzA, and 5yfpH. TM: transmembrane subunit; SU: surface subunit. (F) Tertiary structure of the NHR-CHR trimeric domain of Env38 TM subunit predicted by SWISS-MODEL program with crystal structures of human Syncytin 1 in post-fusion conformation (SMTL ID: 6rx1.1) as a model. A homotrimer with a fusion core structure was formed by three NHR-CHR domains in which three helical NHR peptides formed a central core and three helical CHR peptides packed into the grooves on the surface of the central core. |
|
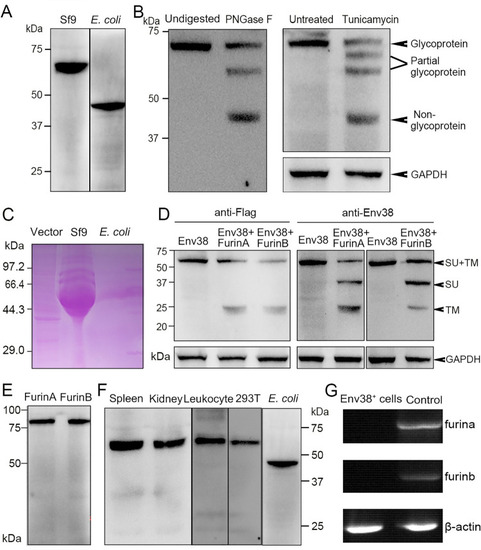
Examination on the expression, glycosylation and proteolysis of Env38 protein and activity of anti-Env38 Ab.
(A) Western blot analysis for the expression of Env38 protein with mouse anti-His Ab (1:5,000). The Env38 proteins expressed in eukaryotic Sf9 cells and prokaryotic E. coli cells were determined to be approximately 60 kDa and 48 kDa, respectively. (B) Western blot analysis for N-glycosylation of Env38 protein with treatment of PNGase F and tunicamycin, in which the Env38 protein derived from Sf9 cells was treated with PNGase F (500 U/ml), and the Env38-expressing HEK293T cells transfected with pcDNA3.1-Flag-Env38-LTR (0.6 μg/ml) was treated with tunicamycin (5 μg/ml). The Env38 protein was separated by 12% SDS-PAGE. Env38 protein derived from Sf9 cells without digestion and HEK293T cells without treatment were served as controls. GAPDH was used as a loading control. (C) Detection of the glycosylation of Env38 protein by PAS reaction. The Env38 proteins were expressed in Sf9 cells or E. coli cells, and separated by 12% SDS-PAGE followed by staining with Schiff reagent. Env38 protein derived from Sf9 cells exhibited a strong positive staining, while Env38 protein derived from E. coli cells showed no staining. (D) Western blot analysis for the detection of enzymatic cleavage of Env38 protein by Furins. Coexpression of Env38 protein (with a Flag-tag at C-terminus) and FurinA or FurinB in HEK293T cells resulted in cleavage of Env38 with two protein bands corresponding to the intact Env38 protein (60 kDa) and TM subunit (25 kDa) when detected with anti-Flag Ab, and three protein bands corresponding to the intact Env38 protein (60 kDa), SU subunit (35 kDa) and TM subunit (25 kDa) when detected with mouse anti-Env38 Ab. GAPDH was used as a loading control. (E) Western blot analysis for the expression of FurinA and FurinB proteins in HEK293T cells. (F) Western blot analysis for the reactivity and specificity of mouse anti-Env38 polyclonal Ab. The anti-Env38 Ab clearly combined with the target natural Env38 proteins derived from spleen, head kidney and leukocytes, as well as recombinant Env38 proteins from HEK293T and E. coli cells with molecular weights of 60 kDa and 48 kDa, respectively. (G) Detection of the transcriptional expression of furina and furinb genes in Env38+ cells sorted from leukocytes from spleen, head kidney and peripheral blood of zebrafish stimulated with SVCV (105 TCID50) using RT-PCR. Total leukocytes were used as a control. |
|
Subcellular localization analysis of Env38 in HEK293T and EPC cells.
(A-C) HEK293T cells were transfected with the recombinant expression plasmid of pEGFPN1-Env38 (0.6 μg/ml) for 48 h, and then fixed and stained with the cell membrane probe DiI (A), ER-tracker (B) or Golgi-tracker (C) and nuclei probe DAPI. (D and E) EPC cells were transfected with the recombinant expression plasmid of pEGFPN1-Env38 (0.6 μg/ml) for 24 h and stimulated with SVCV (100 TCID50) (E) or not (D) for another 24 h, and then fixed and stained with the cell membrane probe DiI and nuclei probe DAPI. The blue, green, and red fluorescence showed DAPI-labeled nuclei, Env38–EGFP fusion protein, and DiI–labeled cell membrane or ER-tracker–labeled ER or Golgi-tracker–labeled Golgi apparatus. Fluorescence images were captured using Laser scanning confocal microscope (FV3000) with 60 × oil glass. |
|
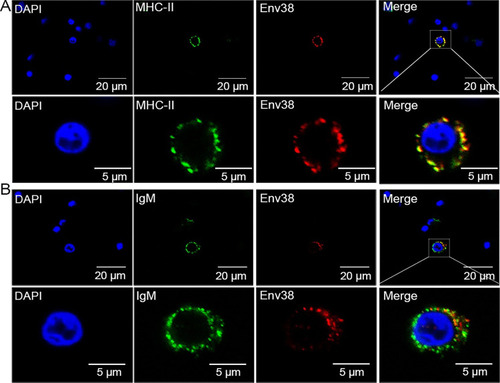
Double immunofluorescence staining of MHC-II+Env38+ APCs in leukocytes of zebrafish.
The leukocytes were sorted from spleen, head kidney and peripheral blood of zebrafish stimulated with SVCV (105 TCID50). (A) Cells were labled with mouse anti-Env38 and rabbit anti-MHC-IIα Abs, followed by Alexa Fluor 594-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG. (B) Cells were labled with mouse anti-Env38 and rabbit anti-IgM Abs, followed by Alexa Fluor 594-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG. DAPI stain showed the location of the nuclei. Fluorescence images were captured using a Laser scanning confocal microscope (FV3000) with 60 × oil glass. |
|
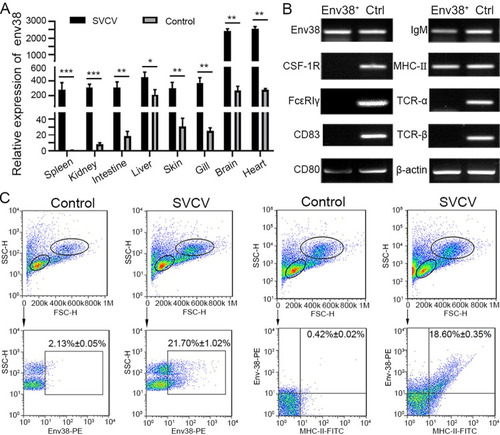
Detection of Env38 in tissues and cells in response to SVCV infection.
(A) RT-qPCR analysis for the expression of env38 gene in spleen, head kidney, intestine, liver, skin, gill, brain and heart tissues of zebrafish stimulated with SVCV (105 TCID50) or mock PBS (control) for 7 days. The expression of env38 mRNA in spleen in PBS treatment was arbitrarily set at 1. Negative control group was treated with mock PBS. Each sample was obtained from at least 10 fish. (B) Cellular distribution of Env38 on leukocytes from spleen, head kidney and peripheral blood of zebrafish stimulated with SVCV (105 TCID50). The Env38+ APCs were sorted from the leukocytes by FCM and qualitatively examined by RT-PCR with a panel of cellular markers of various immune cells, including CSF-1R and FcεRIγ (markers of monocytes/macrophages), CD83 and CD80 (markers of dendritic cells), membrane IgM (markers of B lymphocytes), MHC-II (markers of antigen presenting cells) as well as TCR-α and TCR-β (markers of T lymphocytes). Total leukocyte sample was used as a control. (C) Flow cytometry analysis of the percentages of Env38+ cells and MHC-II+Env38+ cells in lymphatic and myeloid leukocytes from spleen, head kidney and peripheral blood of zebrafish stimulated with SVCV (105 TCID50) for 7 days. The numbers above the outlined areas indicated the percentage of cells in each group. In control groups, zebrafish were i.p. with the same amount of mock PBS. In the RT-qPCR assay, PCRs were run in combination with the endogenous β-actin control. Error bars represent SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
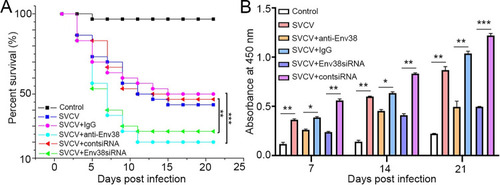
Evaluation on the functional role of Env38 in host antiviral immune defense and IgM antibody production.
(A) Survival rate was calculated in anti-Env38 Ab-mediated blockade and Env38siRNA-LV-mediated knockdown treatment in zebrafish injected with SVCV (105 TCID50) in continuous 21 days. Kaplan-Meier survival curves represented data pooled from three independent experiments. Statistical differences were analyzed by log-rank test. 30 zebrafish were used in each experiment (n = 30). (B) ELISA for kinetic production analysis of the anti-SVCV antibody (IgM) in serum of zebrafish with treatments of Ab-mediated blockade and siRNA-mediated knockdown of Env38 under challenge of SVCV (105 TCID50) after 7, 14 and 21 days. Error bars represented SEM. Isotype IgG-treated groups and scrambled siRNA-LV treated groups served as nonrelated control groups (n = 30). Negative control groups received PBS treatment. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
Evaluation on the functional role of Env38 in APC-initiated Ag-specific CD4+ T cell activation.
(A and B) In vitro examination on the proliferation (A) and activation (B) of CD4+ TSVCV cells isolated from zebrafish stimulated with inactivated SVCV (106 TCID50) and impaired by blockade of Env38 on APCs with anti-Env38 Ab (5 μg/ml) as determined by the CFSE dilution through FCM analysis and the transcriptional expression of cd154 and lck genes through RT-qPCR. (C and D) In vivo examination on the activation of CD4+ T cells stimulated with SVCV (105 TCID50) and impaired by blockade or knockdown of Env38 with i.p administering anti-Env38 Ab (10 μg/fish) or Env38siRNA-LV (1×106 TU/fish) as determined by the percentage of CD4+CD154+ T cells with mouse anti-CD154 (1:500) and rabbit anti-CD4 (1:500) in leukocytes from spleen, head kidney and peripheral blood through FCM analysis (C) and the transcriptional expression of cd154 and lck genes through RT-qPCR (D). (E) In vivo examination on the proliferation of CD4+ T cells of zebrafish stimulated with SVCV (105 TCID50) and impaired by blockade or knockdown of Env38 with i.p administering anti-Env38 Ab (10 μg/fish) or Env38siRNA-LV (1×106 TU/fish) as determined by the percentage of CD4+EdU+ T cells with rabbit anti-CD4 (1:500) in leukocytes from spleen, head kidney and peripheral blood as determined by FCM analysis. Nonrelated control groups were administered with isotype IgG or scrambled siRNA-LV. Negative control groups received mock PBS. The data presented in each block diagram indicated the average percentage of CD4+CD154+ T cells or CD4+EdU+ T cells in each treatment group. RT-qPCRs were run in combination with the endogenous β-actin control. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
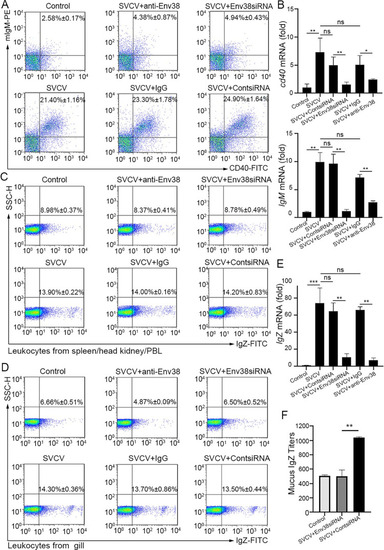
Evaluation on the functional role of Env38 in CD4+ T cell-initiated B cell activation.
(A and B) In vivo examination on the activation of IgM+ B cells of zebrafish stimulated with SVCV (105 TCID50) and impaired by blockade or knockdown of Env38 with i.p administering anti-Env38 Ab (10 μg/fish) or Env38siRNA-LV (1×106 TU/fish) as determined by the percentage of mIgM+CD40+ B cells with mouse anti-mIgM (1:2,000) and rabbit anti-CD40 (1:500) through FCM analysis (A) and the transcriptional expression levels of cd40 and IgM genes through RT-qPCR (B). (C-F) In vivo examination on the activation of IgZ+ B cells of zebrafish stimulated with SVCV (105 TCID50) and impaired by blockade or knockdown of Env38 with i.p administering anti-Env38 Ab (10 μg/fish) or Env38siRNA-LV(1×106 TU/fish) as determined by the percentage of IgZ+ B with rabbit anti-IgZ (1:500) in leukocytes from spleen, head kidney and peripheral blood (C) and from gill (D) through FCM analysis, the transcriptional expression of IgZ genes via RT-qPCR (E) and the mucus IgZ antibody production by ELISA (F). Nonrelated control groups were administered with isotype IgG or scrambled siRNA-LV. Negative control groups received mock PBS. The data presented in each block diagram indicated the average percentage of IgM+CD40+B or IgZ+ B cells in each treatment group. RT-qPCRs were run in combination with the endogenous β-actin control. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
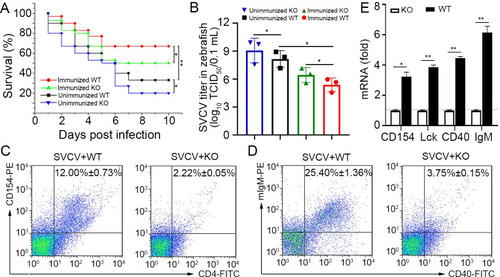
Verification of the functional role of Env38 in knockout zebrafish.
(A) The functional role of Env38 was verified by the impairment of the antiviral activity of Env38-/- zebrafish after vaccination with formaldehyde-inactivated SVCV vaccine or infection with live SVCV. In both wildtype and knockout zebrafish, the immunized groups were i.p. inoculated with vaccine derived from 0.4% formaldehyde-inactivated SVCV (106 TCID50) and then challenged with SVCV (105 TCID50); whereas the unimmunized groups were challenged with SVCV (105 TCID50) without vaccination. Data points were from three independent experiments, and 30 zebrafish were used in each experiment (n = 30). Differences were analyzed using log-rank test. (B) Viral titer (TCID50) was measured to evaluate the viral load in the wild type and knockout zebrafish with or without vaccinated immunoprotection. Viral samples were prepared from zebrafish at 4th day post SVCV (105 TCID50) infection. The TCID50 was examined in EPC cells by using Reed-Muench method. Viral titers were log10 transformed prior to statistical analysis. Data points were from three independent experiments, and three zebrafish were used for viral sample preparation in each experiment (n = 3). (C and D) The degree of CD4+ T and IgM+ B cell activation between WT and KO zebrafish under SVCV (105 TCID50) stimulation was represented by the percentage of CD4+CD154+ T and IgM+CD40+ B cells with mouse anti-CD154 (1:500), rabbit anti-CD4 (1:500) (C) and mouse anti-mIgM (1:2,000), rabbit anti-CD40 (1:500) (D) through determination with FCM analysis. The data presented in each block diagram indicated the average percentage of CD4+CD154+ T and IgM+CD40+ B cells in WT and KO zebrafish. (E) The degree of CD4+ T or IgM+B cell activation between WT and KO zebrafish under SVCV (105 TCID50) stimulation was represented by the transcriptional levels of cd154, lck, cd40 and IgM through RT-qPCR. The RT-qPCRs were run in combination with the endogenous β-actin control. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
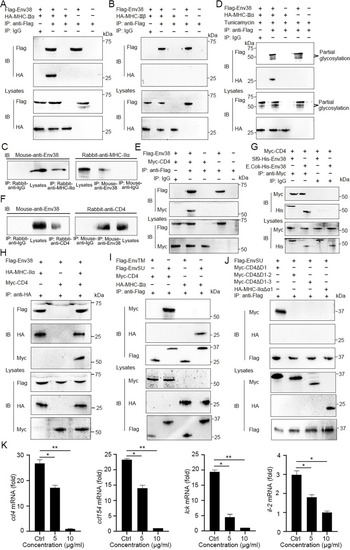
Examination on the association of Env38 with MHC-II and CD4 proteins and their domain interactions.
(A and B) Examination on the interaction between Env38 and MHC-IIα or MHC-IIβ, in which the Flag-tagged Env38 and HA-tagged MHC-IIα (A) or HA-tagged MHC-IIβ (B) were coexpressed in HEK293T cells, followed by immunoprecipitation with rabbit anti-Flag or rabbit IgG (negative control). Western blot was performed with mouse anti-Flag or anti-HA Ab. (C) Examination on the interaction between leukocyte Env38 and MHC-IIα proteins of zebrafish under SVCV stimulation. The lysates were immunoprecipitated with rabbit anti-MHC-IIα or rabbit IgG. Western blot was performed with mouse anti-Env38 Ab. Similarly, the lysates were immunoprecipitated with mouse anti-Env38 or mouse IgG, and Western blot was performed with rabbit anti-MHC-IIα Ab. (D) Examination on the effect of glycosylation on the interaction between Env38 and MHC-IIα. The Flag-tagged Env38 and HA-tagged MHC-IIα were coexpressed in HEK293T cells under treatment with tunicamycin, followed by immunoprecipitation with rabbit anti-Flag or rabbit IgG. Western blot was performed with mouse anti-Flag or anti-HA Ab. (E) Examination on the interaction between Env38 and CD4, in which the Flag-tagged Env38 and the Myc-tagged CD4 were coexpressed in HEK293T cells, followed by immunoprecipitation with rabbit anti-Flag or rabbit IgG. Western blot was performed with the mouse anti-Flag or anti-Myc Ab. (F) Examination on the interaction between leukocyte Env38 and CD4 proteins of zebrafish under SVCV stimulation. The lysates were immunoprecipitated with rabbit anti-CD4 or rabbit IgG. Western blot was performed with mouse anti-Env38 Ab. Similarly, the lysates were immunoprecipitated with mouse anti-Env38 or mouse IgG, and Western blot was performed with rabbit anti-CD4 Ab. (G) Examination on the interactions between Env38 and CD4 proteins, in which the His-tagged Env38 proteins were purified from Sf9 or E. coli cells and the Myc-tagged CD4 was expressed in HEK293T cells, followed by immunoprecipitation with rabbit anti-Myc or rabbit IgG. Western blot analysis was performed with the mouse anti-Myc or anti-His Ab. (H) Examination on the interactions among Env38, MHC-IIα and CD4, in which the Flag-tagged Env38, HA-tagged MHC-IIα and Myc-tagged CD4 were coexpressed in HEK293T cells, followed by immunoprecipitation with rabbit anti-HA Ab. Western blot was performed with mouse anti-Flag, anti-HA or anti-Myc Ab. (I) Examination on the interactions among EnvTM, EnvSU, CD4 and MHC-IIα, in which the Flag-tagged EnvTM, Flag-tagged EnvSU, Myc-tagged CD4 and HA-tagged MHC-IIα were coexpressed in HEK293T cells in different combinations, followed by immunoprecipitation with rabbit anti-Flag Ab. Western blot was performed with mouse anti-Myc, anti-HA, or anti-Flag Ab. (J) Examination on the interactions among EnvSU, CD4ΔD1, CD4ΔD1-2, CD4ΔD1-3 and MHC-IIαΔα1, in which the Flag-tagged EnvSU, Myc-tagged CD4ΔD1, Myc-tagged CD4ΔD1-2, Myc-tagged CD4ΔD1-3 and HA-tagged MHC-IIαΔα1 were coexpressed in HEK293T cells in different combinations, followed by immunoprecipitation with rabbit anti-Flag Ab. Western blot was performed with mouse anti-Myc, anti-HA, or anti-Flag Ab. The same lysates were simultaneously immunoblotted to monitor the expression of CD4ΔD1, CD4ΔD1-2, CD4ΔD1-3, MHC-IIαΔα1, and EnvSU. In the Co-IP assays, the same lysates were simultaneously immunoblotted to monitor the expression of proteins in input. (K) Functional evaluation on the association among Env38, MHC-IIα and CD4 proteins distributed on MHC-II+Env38+APCs and CD4+ T cells by examining the competitive inhibition of CD4+ T cell activation by using a soluble D2 domain protein of CD4 (sCD4-D2, 5 μg/ml and 10 μg/ml). The activation of CD4+ T cells was determined by the transcriptional expression levels of cd4, cd154, lck and il-2 genes through RT-qPCR. The RT-qPCRs were run in combination with the endogenous β-actin control. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |
|
A proposed model of the potential TCR-pMHC-II-CD4-Env38 complex.
The TCR-pMHC-II-CD4-Env38 complex formed in the immune synapse structure between MHC+Env38+ APC and CD4+ T cell during APC-initiated CD4+ T cell activation. Env38 located on the surface of MHCII+ APC in an integral form without undergoing cleavage between the transmembrane subunit (TM) and the surface subunit (SU). Env38 was associated with MHC-IIα and CD4 proteins, in which the SU subunit of Env38, the second immunoglobin domain of CD4 (CD4-D2) and the first α1 domain of MHC-IIα chain (MHC-IIα1) contribute to the association among Env38, CD4 and MHC-IIα proteins. In particular, Env38 specifically binds to CD4-D2 and MHC-IIα1 domains without intervention with the CD4-D1 and MHC-IIβ-β2 as well as CD4-D1 and MHC-IIα2 domains, two previously known reciprocal connectors contributing to the formation of the pMHC-TCR-CD4 complex referred as the central supramolecular activating complex (c-SMAC) in the immunological synapse. Hence, Env38 promoted the formation and/or stabilization of pMHC-TCR-CD4 complex via cross-linking MHC-II and CD4 molecules between the MHCII+ APCs and CD4+ T cells. The red circles represented the interaction domains among SU, CD4-D2 and MHC-IIα1. |
|
Examination on the stimulatory role of zebrafish IFNφ1/IFNφ2 in Env38 production.
(A-C) Examination on the stimulatory role of zebrafish IFNφ1 and IFNφ2 in Env38 expression by in vivo administering recombinant IFNφ1 and IFNφ2 proteins. The stimulatory effect was examined by the increased percentage of Env38+ cells and the upregulated transcriptional expression of env38, isg15 and viperin genes in leukocytes from spleen, head kidney and peripheral blood of zebrafish i.p. injected with proportional dosage (2.5 μg, 5 μg and 10 μg) of the IFNφ1 (A) or IFNφ2 (B) through FCM analysis (A and B) and RT-qPCR (C), respectively. Zebrafish i.p. injected with mock PBS was used as control. (D and E) Examination on the stimulatory role of zebrafish IFNφ1 in Env38 expression by in vivo neutralization and rescue assays via administering mouse anti-IFNφ1 Ab (10 μg/fish) and recombinant IFNφ1 (10 μg/fish) back into the IFNφ1-neutralized zebrafish. The stimulatory effect was examined by the percentage of Env38+ cells (D) and the expression level of env38, isg15 and viperin genes (E) in leukocytes from spleen, head kidney and peripheral blood of zebrafish in each group under stimulation with SVCV (105 TCID50). Zebrafish in the control groups were treated with isotype mouse IgG and rescued with mock PBS. (F) Western blot analysis of the recombinant IFNφ1 and IFNφ2 proteins with anti-Flag Ab (1:5,000) from supernatant of HEK293T cells transfected with the pcDNA3.1-His-Flag-IFNφ1 and pcDNA3.1-His-Flag-IFNφ2 recombinant constructs or an empty control construct. In the FCM analysis, different treatments were presented at the top of each block diagram, and the data presented in each block diagram indicated the average percentage of Env38+ cells in each treatment group. RT-qPCRs were run in combination with the endogenous β-actin control. Error bars represented SEM. (*p < 0.05; **p < 0.01; ***p < 0.001; ns, no significant difference). |