- Title
-
Identification of Novel Coloboma Candidate Genes through Conserved Gene Expression Analyses across Four Vertebrate Species
- Authors
- Trejo-Reveles, V., Owen, N., Ching Chan, B.H., Toms, M., Schoenebeck, J.J., Moosajee, M., Rainger, J., Genomics England Research Consortium
- Source
- Full text @ Biomolecules
|
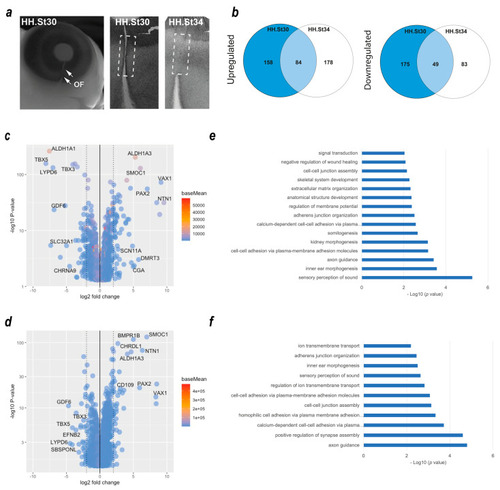
Transcriptome profiling of chicken optic fissure closure. ( |
|
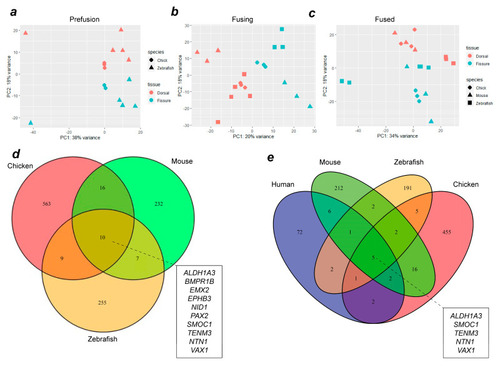
Combined transcriptome analysis across vertebrate species reveals common gene expression signatures during OFC. PCA analyses for aligned transcriptomes for chicks and zebrafish ( |
|
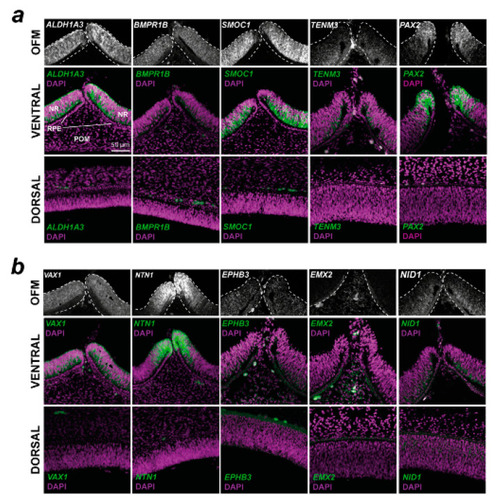
Spatial gene expression analysis for 10 conserved fissure-enriched genes in the developing chick eye. Fluorescent in situ hybridisation for ( |
|
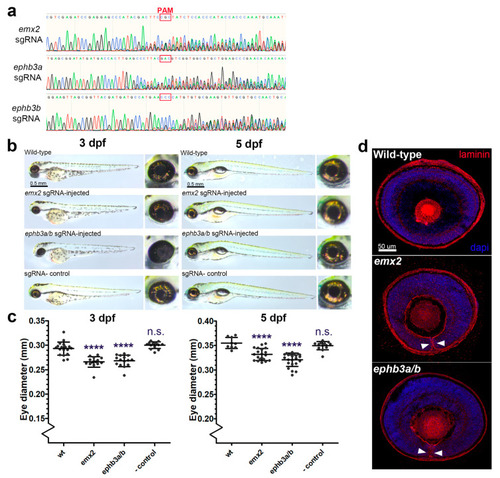
Targeted gene disruption of OFC novel candidates using CRISPR/Cas9 gene targeting in zebrafish embryos. ( EXPRESSION / LABELING:
PHENOTYPE:
|