- Title
-
Analyses of binding partners and functional domains for the developmentally essential protein Hmx3a/HMX3
- Authors
- Haws, W., England, S., Grieb, G., Susana, G., Hernandez, S., Mirer, H., Lewis, K.
- Source
- Full text @ Sci. Rep.
|
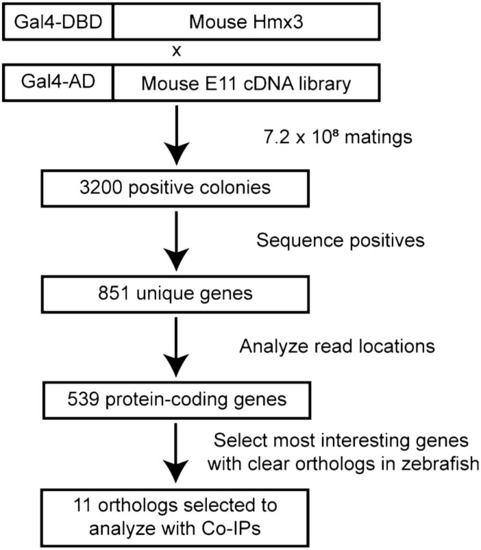
Isolation of novel binding partners of HMX3 with yeast two-hybrid screening. Top: Schematic representation of bait and prey constructs used in screening. Full-length mouse HMX3 fused to the Gal4 DNA-binding domain (DBD) was used as bait. A library of stage E11 mouse cDNAs fused to the Gal4 activating domain (AD) was used as prey. 7.2 × 108 solid plate matings were performed, yielding 3200 positive colonies. Positive prey inserts were segregated and sequenced and reads were aligned to the mouse genome, identifying 851 unique genes. After removing non-coding genes and those for which only untranslated region (UTR) or intronic sequences were recovered, 539 protein-coding sequences remained. Zebrafish orthologs of the most functionally interesting genes were selected for further analysis with Co-IPs. |
|
Zebrafish Prmt2, Azin1b, Hmgn3, Hmgb1a, and the WDR domain of Tle3b bind full-length zebrafish Hmx3a in Co-IPs and have varying interaction profiles with proteins encoded by |
|
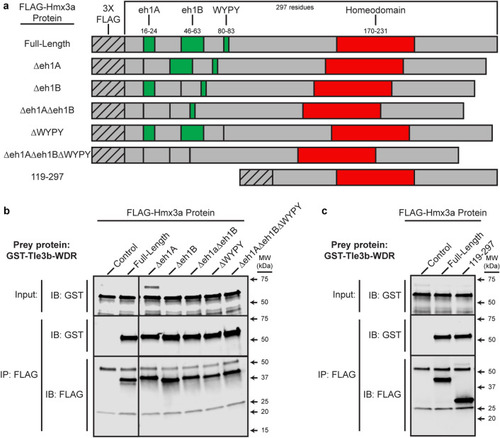
The WDR domain of Tle3b binds to the C-terminus of Hmx3a but not to any canonical Tle interaction motifs. (a) Schematics of FLAG-tagged Hmx3a constructs used in these experiments. eh1A, eh1B, and WYPY indicate predicted interaction motifs for Tle3b-WDR. Numbers (top) indicate residues encompassing annotated domains or (left-hand side, bottom row) included in the C-terminal construct. (b, c) The WDR domain of Tle3b was expressed as a GST-fusion protein, purified, and incubated with either a FLAG-Hmx3a-expressing embryo lysate or a stage-matched control lysate expressing no recombinant Hmx3a. Proteins were immunoprecipitated with an anti-FLAG antibody and separated by SDS-PAGE followed by transfer to a membrane and immunoblotting with anti-FLAG or anti-GST antibody. Top images in each panel show anti-GST immunoblot (IB) of 4% of input. Middle and bottom images in each panel show blots for GST and FLAG, respectively, of the immunoprecipitate (IB). MW is shown on the right-hand side. (b) Co-IPs with putative interaction motif deletion constructs. Blot images on each side of respective rows were from the same membrane and were processed identically for brightness/contrast. (c) Co-IP with carboxy-terminal Hmx3a construct. FLAG antibody heavy and light chains appear at just above 50 kDa and at 25 kDa, respectively in IP:FLAG;IB:FLAG fractions. Note that though it lacks 9 amino acids vs full-length Hmx3a, FLAG-Hmx3a-Δeh1A runs slightly above full-length Hmx3a (panel (b) IP:FLAG;IB:FLAG). A non-specific band, likely corresponding to the yolk protein Vitellogenin, is also visible above GST-Tle3b-WDR in the anti-GST input fraction of Δeh1A and, very slightly, in the control in panel (b). n = at least 2 for all experiments. Original blots are presented in Supplementary Fig. S3. |