- Title
-
Genomic Structure, Protein Character, Phylogenic Implication, and Embryonic Expression Pattern of a Zebrafish New Member of Zinc Finger BED-Type Gene Family
- Authors
- Zeng, C.W., Sheu, J.C., Tsai, H.J.
- Source
- Full text @ Genes (Basel)
|
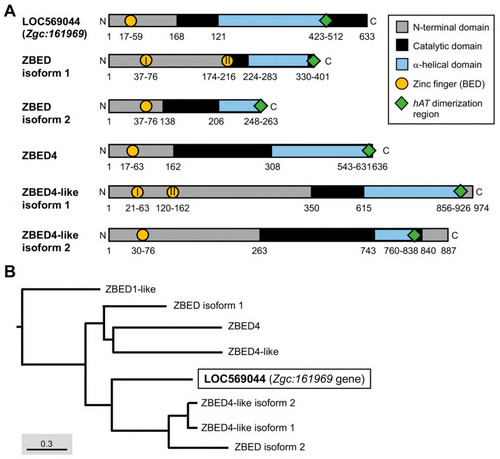
The essential domains of the ZBED protein family in zebrafish and their phylogenetic tree. (A) Different domains were labeled by various colors as indicated. Arabic numbers represent the position of amino acid residues located at each domain, while Roman numerals presented in circles indicate the number of zinc finger BED domains existing in ZBED proteins. (B) Phylogenetic tree for ZBED proteins in zebrafish (transcript ID in Table 1). Three monophyletic clades indicated by different colors are categorized. The uncharacterized LOC569044 protein encoded by the Zgc:161969 gene is marked by a black box. |
|
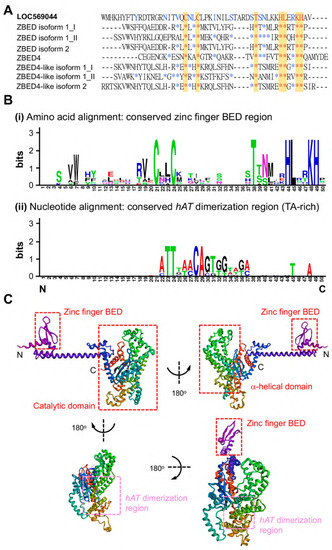
Comparison of the amino acid sequences of ZBED domain between LOC569044 protein and other zebrafish ZBED proteins. (A) The alignment of amino acids of BED domains among zebrafish ZBED proteins. The DNA-binding BED domain was characterized by the signature Cx2CxnHx3-5H (xn indicates a variable spacer; yellow color). Blue color indicates that at least one amino acid residue among zebrafish ZBED proteins was identical to that of LOC569044 protein, while the red color indicates amino acid residue in all zebrafish ZBED proteins entirely identical to that of the LOC569044 protein. *: indicates that the amino acid residue is identical among all species examined. (B) Using the WebLogos (version 2.8.2) to analyze the consensus residues of amino acid and nucleic acid of conserved zinc finger BED domain (i) and hAT dimerization region (ii), respectively. The larger size of alphabetic letters represent a higher degree of consensus. (C) A predicted protein structure of LOC569044 protein. Four views of the predicated protein structures were demonstrated by a 180-degree rotation. Positions of N- and C-termini are labeled. Positions of the three domains are labeled by red boxes, and a hAT dimerization region is labeled by pink box. |
|
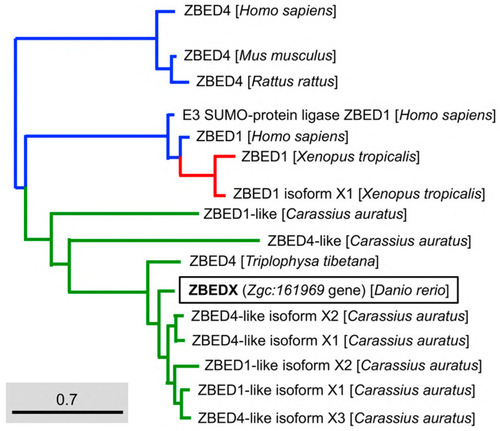
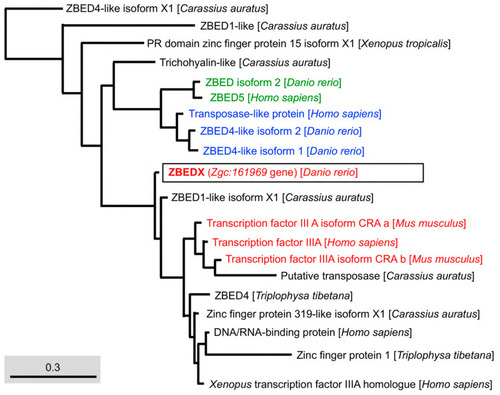
Phylogenetic analysis of the LOC569044 protein and the ZBED proteins found in mammals, amphibians, and bony fish. The full amino acid sequences of ZBED proteins from the known species are human (H. sapiens), mouse (M. musculus), zebrafish (D. rerio), frog (X. tropicalis), goldfish (C. auratus), and stone loach (T. tibetana), which were used to characterize the LOC569044 protein. ZBED proteins from known species were categorized into three main monophyletic clades labeled in different colors (transcript ID in Table 2). The LOC569044 protein marked by a black box was categorized in the same monophyletic group, along with the ZBED of bony fish (in green). The scale bar represents the number of substitutions per site. The blue and red colors indicate the same monophyletic group in the mammals and amphibians in, respectively. |
|
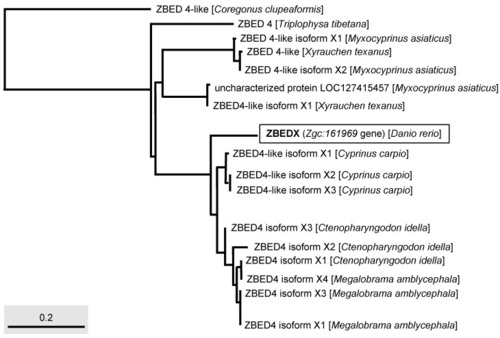
Phylogenetic analysis of the LOC569044 protein and ZBED protein found in multiple bony fishes. The full-length amino acid sequences of ZBED proteins from different species of bony fishes, such as C. clupeaformis, T. tibetana, M. asiaticus, X. texanus, C. carpio, C. idella, and M. amblycephala used to characterize the LOC569044 protein. The LOC569044 protein is marked by a rectangular box. The scale bar represents the number of substitutions per site. |
|
Phylogenetic analysis of the LOC569044 protein and the ZBED domains found in mammals, amphibians, and bony fish. The full amino acid sequences of ZBED domains in ZBED proteins from the known species are human (H. sapiens), mouse (M. musculus), zebrafish (D. rerio), frog (X. tropicalis), goldfish (C. auratus), and stone loach (T. tibetana), which were used to characterize the LOC569044 protein. The LOC569044 protein is marked by a black box. The proteins labeled in the same color are recognized as a group of proteins that are close in evolution and function of BED domains. Scale bar represents the number of substitutions per site. |
|
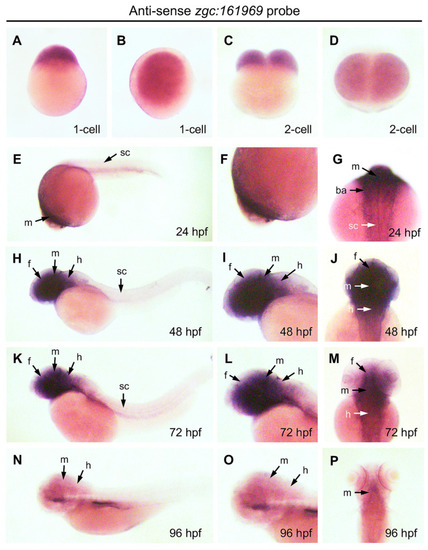
Expression pattern of ZBEDX transcripts during the early developmental stages of zebrafish embryos. Embryos at different stages, as indicated, were collected and hybridized with antisense probe against ZBEDX mRNA using whole-mount in situ hybridization. Panels (A,C,E,H,K,N) are lateral views, while panels (B,D,F,I,J,L,M,O,P) are dorsal views. Panels (E,H,K,N) are lateral views, and the anterior of embryos was placed at the left, while panels (G,J,M,P) are dorsal views, and the anterior of embryos is on the top. At 12 hpf, ZBEDX transcripts were expressed in whole embryo. ZBEDX transcripts were initially apparent at the 1- (A,B) and 2-cell (C,D) stages. At 24–72 hpf, ZBEDX transcripts were expressed in forebrain (f), midbrain (m), hindbrain (h), retina (r), spinal cord (sc), and branchial arches (ba) (E–M). At 96-hpf, ZBEDX transcripts were expressed in midbrain (m) and hindbrain (h) (N–P). |