- Title
-
Regeneration and developmental enhancers are differentially compatible with minimal promoters
- Authors
- Begeman, I.J., Emery, B., Kurth, A., Kang, J.
- Source
- Full text @ Dev. Biol.
|
E1b, P0.8, and P2 drive minimal leaky expression in uninjured tissues. (A) Experimental design. Transgenic constructs were injected into one-cell-stage wild-type or Tg(cmlc2:mCherry-NTR) embryos, and EGFP expression was observed at 3 dpf. (B) 3-dpf F0 uninjured hearts, eyes, and fin folds. Arrows indicate EGFP expression. Eyes are demarcated by dashed lines. (C, D) Tables of larvae displaying EGFP expression in each tissue for each promoter alone (C) or when paired with LEN (D). Scale bars: 100 μm. |
|
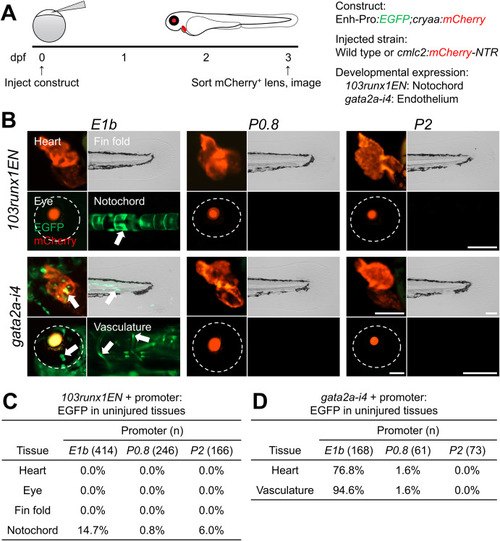
Developmental enhancer activity is preferentially compatible with E1b. (A) Experimental design. Transgenic constructs were injected into one-cell-stage wild-type or Tg(cmlc2:mCherry-NTR) embryos, and EGFP expression was observed at 3 dpf. (B) 3-dpf F0 hearts, eyes, fin folds, notochords, and vasculature in uninjured larvae. Arrows indicate EGFP expression. Eyes are demarcated by dashed lines. (C, D) Tables of larvae displaying EGFP expression in each tissue for each promoter when paired with 103runx1EN (C) or gata2a-i4 (D). Scale bars: 100 μm. |
|
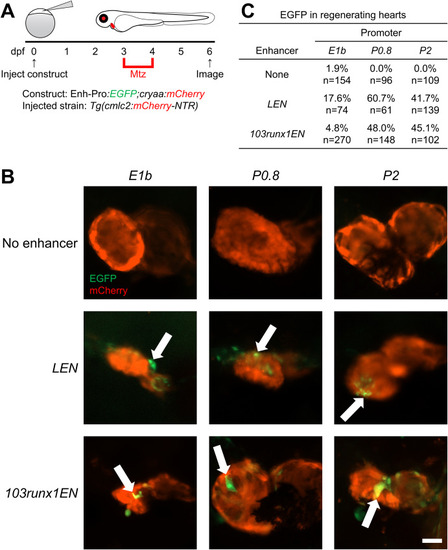
Regeneration enhancer activity in hearts is preferentially compatible with P0.8 and P2. (A) Experimental design. Transgenic constructs were injected into one-cell-stage Tg(cmlc2:mCherry-NTR) embryos, larvae were treated with Mtz for 24 h starting at 3 dpf to ablate cardiomyocytes, and EGFP expression was observed at 6 dpf. (B) 6-dpf F0 regenerating hearts following Mtz-induced cardiomyocyte ablation. Arrows indicate EGFP expression. (C) Table of larvae displaying cardiac EGFP expression for each promoter alone or when paired with LEN or 103runx1EN. Scale bar: 50 μm. |
|
Injury-induced activity of enhancer-promoter constructs in amputated fin folds. (A) Experimental design. Transgenic constructs were injected into one-cell-stage wild-type embryos, fin folds were amputated at 4 dpf, and EGFP expression was observed at 6 dpf. (B) 6-dpf F0 regenerating fin folds following injury. Arrows indicate EGFP expression. (C) Table of larvae displaying injury-induced fin fold EGFP expression for each promoter alone or when paired with LEN or 103runx1EN. Scale bar: 100 μm. |
|
Activity of minimal promoters and regeneration enhancers in injured eyes. (A) Experimental design. Transgenic constructs were injected into one-cell-stage wild-type embryos, eyes were injured at 4 dpf, and EGFP expression was observed at 6 dpf. (B) 6-dpf F0 regenerating eyes following injury. Arrows indicate EGFP expression. (C) Table of larvae displaying eye EGFP expression for each promoter alone or when paired with LEN or 103runx1EN. Scale bar: 100 μm. |
|
Adding downstream elements to the P0.8 promoter increases developmental enhancer activity. (A) Structure of the ZSP synthetic promoter. (B–E) 3-dpf F0 uninjured heart, eye, fin fold, notochord, and vasculature of ZSP (B), LEN-ZSP (C), 103runx1EN-ZSP (D), and gata2a-i4-ZSP (E) larvae. Arrows indicate EGFP expression. Eyes are demarcated by dashed lines. The percentage of EGFP+ larvae were compared between larvae harboring each ZSP construct and each corresponding P0.8 construct. (F–H) 6-dpf F0 regenerating hearts, eyes, and fin folds following injury in ZSP (F), LEN-ZSP (G), and 103runx1EN-ZSP (H) larvae. (I) Table of larvae displaying EGFP expression in each tissue for ZSP alone or when paired with LEN or 103runx1EN. Scale bars: 100 μm ∗∗∗∗, p < 0.0001. |
Reprinted from Developmental Biology, 492, Begeman, I.J., Emery, B., Kurth, A., Kang, J., Regeneration and developmental enhancers are differentially compatible with minimal promoters, 47-58, Copyright (2022) with permission from Elsevier. Full text @ Dev. Biol.