- Title
-
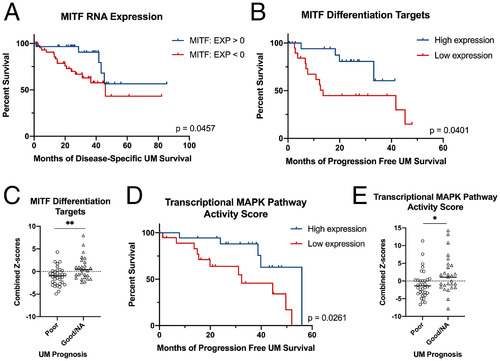
MITF deficiency accelerates GNAQ-driven uveal melanoma
- Authors
- Phelps, G.B., Hagen, H.R., Amsterdam, A., Lees, J.A.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
|
|
EXPRESSION / LABELING:
|
|
|
|
|
|
|