- Title
-
Ruvbl2 Suppresses Cardiomyocyte Proliferation During Zebrafish Heart Development and Regeneration
- Authors
- Sharpe, M., González-Rosa, J.M., Wranitz, F., Jeffrey, S., Copenhaver, K., Burns, C.G., Burns, C.E.
- Source
- Full text @ Front Cell Dev Biol
|
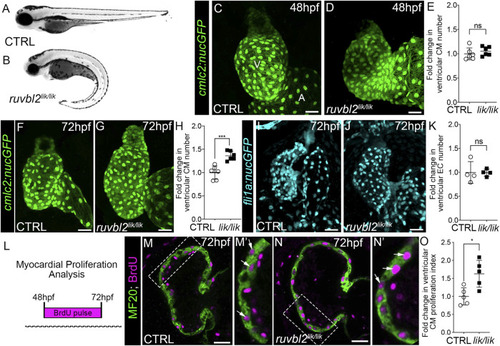
ruvbl2 lik mutant hearts show increased cardiomyocyte proliferation. Brightfield images of wild-type (A) and ruvbl2 lik/lik (B) embryos at 72 hpf. Anterior left, lateral view. (C,D) Confocal projections of fluorescent cardiomyocyte nuclei in hearts of 48 hpf control (CTRL) sibling (C; n = 6) and ruvbl2 lik/lik (D, n = 6) Tg (cmlc2:nucGFP) embryos V = ventricle; A = atrium. (E) Quantification of fold change in ventricular cardiomyocyte number in 48 hpf CTRL and ruvbl2 lik/lik hearts. Means ± s.d are shown. Not significant (ns); p > 0.05. (F,G) Confocal projections of fluorescent cardiomyocyte nuclei in hearts of 72 hpf CTRL (F; n = 6) and ruvbl2 lik/lik (G; n = 6) Tg (cmlc2:nucGFP) embryos. (H) Quantification of fold change in ventricular cardiomyocyte number in 72 hpf CTRL and ruvbl2 lik/lik hearts. Means ± s.d are shown. ***p < 0.0005. (I,J) Confocal projections of fluorescent endocardial nuclei in hearts of 72 hpf CTRL (I; n = 4) and ruvbl2 lik/lik (J; n = 4) Tg (fli1a:nucGFP) embryos. (K) Quantification of fold change in ventricular endocardial cell number in 72 hpf CTRL and ruvbl2 lik/lik hearts. Means ± s.d are shown. Not significant (ns); p > 0.05. (L) Schematic representing the myocardial proliferation assay by BrdU pulse between 48 and 72 hpf. (M–N′) Single plane confocal images of CTRL (M; n = 5) and ruvbl2 lik/lik (N; n = 5) hearts double immunostained to detect cycling (BrdU+) cardiomyocytes (MF20+) at 72 hpf. Boxed regions in (M,N) are magnified in (M′,N′) with white arrows highlighting double positive cells. (O) Quantification of cardiomyocyte proliferation indices in CTRL and ruvbl2 lik/lik ventricles. Means ± s.d are shown. *p < 0.05. Scale bars: 25 μm. |
|
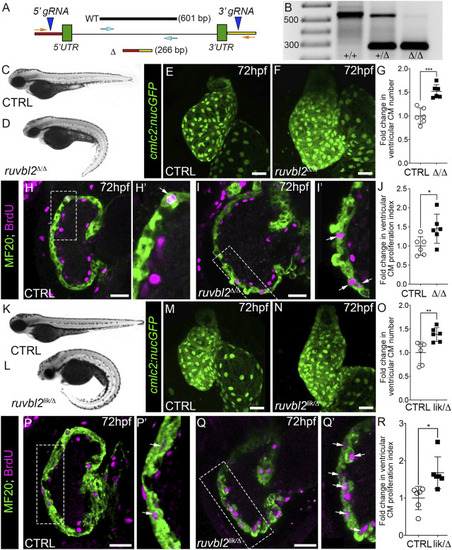
Ruvbl2 suppresses cardiomyocyte proliferation during embryonic heart growth. (A) Schematic of the ruvbl2 locus deletion (ruvbl2 Δ ) targeting strategy by CRISPR-Cas9 genome editing. gRNAs (blue arrowheads) were designed that target genomic regions outside of the ruvbl2 5′ and 3′ UTR. Location of PCR primers used to detect the wild-type (WT; turquoise arrows) and mutant (Δ; orange arrows) alleles. (B) Image of a DNA agarose gel showing amplicons for the wild-type (+) and mutant (Δ) alleles. (C,D) and (K,L) Brightfield images of wild-type (C,K), ruvbl2 Δ/Δ (D), ruvbl2 lik/Δ (L) embryos at 72 hpf. Anterior left, lateral view. (E,F) and (M,N) Confocal projections of fluorescent cardiomyocyte nuclei in hearts of 72 hpf CTRL (F; n = 6 and M; n = 6), ruvbl2 Δ/Δ (G; n = 6), and ruvbl2 lik/Δ (N; n = 6) Tg (cmlc2:nucGFP) embryos. (G,O) Quantification of fold change in ventricular cardiomyocyte number in 72 hpf CTRL and ruvbl2 Δ/Δ (G) or CTRL and ruvbl2 lik/Δ (O) hearts. Means ± s.d are shown. ***p < 0.001; **p < 0.01. (H–I’,P–Q′) Single plane confocal images of CTRL (H; n = 6 and P; n = 6), ruvbl2 Δ/Δ (N; n = 6), and ruvbl2 lik/Δ (Q; n = 6) hearts double immunostained to detect cycling (BrdU+) cardiomyocytes (MF20+) at 72 hpf. Boxed regions in (H,I,P,Q) are magnified in (H′,I′,P′,Q′) with white arrows highlighting double positive cells. (J,R) Quantification of cardiomyocyte proliferation indices in CTRL and ruvbl2 Δ/Δ (J) or CTRL and ruvbl2 lik/Δ (R) ventricles. Means ± s.d are shown. *p < 0.05. Scale bars: 25 μm. |
|
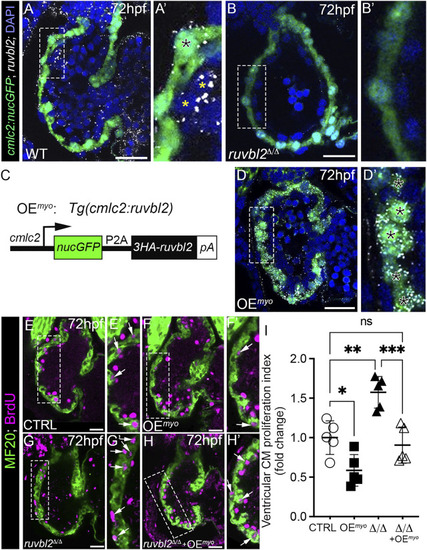
Myocardial expression of Ruvbl2 rescues cardiomyocyte hyperproliferation in ruvbl2 Δ/Δ mutants. (A–B′) Confocal single plane optical sections of endogenous ruvbl2 expression (white signal) relative to myocardium (green signal) processed by RNAscope fluorescent in situ hybridization and immunohistochemistry for GFP in wild-type (A, A′; n = 4) or ruvbl2 Δ/Δ (B, B′; n = 4) Tg (cmlc2:nucGFP) embryos with DAPI nuclei counterstaining (blue signal). Boxed regions in A and B are shown at higher magnifications in A′ and B′, respectively. The black asterix in A′ denotes myocardial ruvbl2 expression, while yellow astrices mark presumptive endocardial ruvbl2 expression. (C) Schematic of the transgene generated to achieve constitutive ruvbl2 overexpression in the myocardium (OEmyo). (D) Confocal single plane optical section of the ruvbl2 expression domain (white signal) relative to myocardium (green signal) processed by RNAscope fluorescent in situ hybridization and immunohistochemistry for GFP in Tg (cmlc2:ruvbl2) Tg (cmlc2:nucGFP) double transgenic embryos (n = 4) with DAPI nuclei counterstaining (blue signal). Boxed region in D is shown at higher magnifications in D′. Black asterices in (D′) denote myocardial ruvbl2 expression, which when compared to wildtype (A,A′) is substantially higher. (E–H′) Single plane confocal images of wildtype control (E,E′), OEmyo (F,F′), ruvbl2 Δ/Δ (G,G′), ruvbl2 Δ/Δ with OEmyo (H,H′) hearts double immunostained to detect cycling (BrdU+) cardiomyocytes (MF20+) at 72 hpf. Boxed regions in (E–H) are magnified in (E′–H′) with white arrows highlighting double positive cells. Sample sizes are n = 6 for all groups. (I) Quantification of fold change in cardiomyocyte proliferation indices between cohorts. Means ± s.d are shown. ns; not significant; *p < 0.05, **p < 0.01, ***p < 0.001. Scale bars: 25 μm. |
|
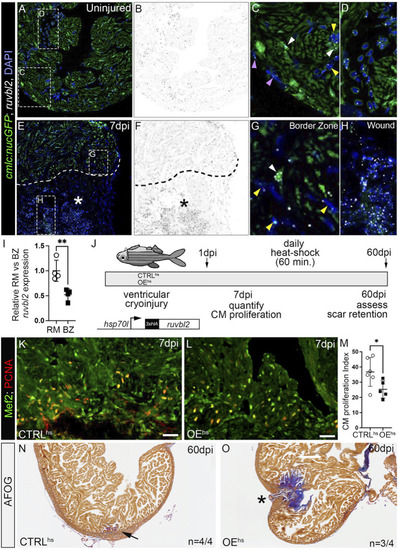
ruvbl2 suppresses cardiomyocyte proliferation during zebrafish cardiac regeneration and leads to scar retention. (A–H) RNAScope in situ hybridization of ruvbl2 transcripts in representative cardiac sections from uninjured or 7 dpi hearts immunostained for GFP and counterstained with DAPI. Ruvbl2 fluorescent signals in (A,E) are pseudocolored to black and white in (B,F). Boxed regions in A and E are shown in magnified views in (C,D,G,H) as indicated. (C,G) Purple arrowhead, epicardial cells; white arrowhead, myocardial cells; yellow arrowhead, endocardial cells. (D) Ruvbl2 expression in circulating cells. (E,F) Dotted line shows the border between the wound (marked with an asterisk) and the spared cardiac tissue. (I) Quantification of relative ruvbl2 expression between the remote myocardium (RM) and border zone (BZ). (J) Schematic of experimental design for regeneration experiments. Adult zebrafish carrying the Tg (hsp70l:ruvbl2) overexpression transgene were given a single heat shock per day following cryoinjury to the ventricle. Experimental endpoints were at 7 dpi and 60 dpi to assay cardiomyocyte proliferation and regeneration, respectively. (K,L) Sections of WT and Tg (hsp70l:ruvbl2) hearts immunostained for Mef2c and PCNA at 7 dpi. (M) Quantification of the cardiomyocyte proliferation index at 7 dpi. (N,O) Representative sections of WT and Tg (hsp70l:ruvbl2) hearts stained with acid fuchsin orange G (AFOG) at 60 dpi. Black arrow highlights regenerated wound area. Black asterisk highlights fibrotic scar tissue with open myocardial wall. 4 of 4 WT hearts regenerated well and 3 of 4 OEhs hearts retained substantial scar tissue as shown. Mean ± SD are shown. *p < 0.05; **p < 0.01. Scale bars: 25 μm. |