- Title
-
The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization
- Authors
- Fujihara, Y., Herberg, S., Blaha, A., Panser, K., Kobayashi, K., Larasati, T., Novatchkova, M., Theussl, H.C., Olszanska, O., Ikawa, M., Pauli, A.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
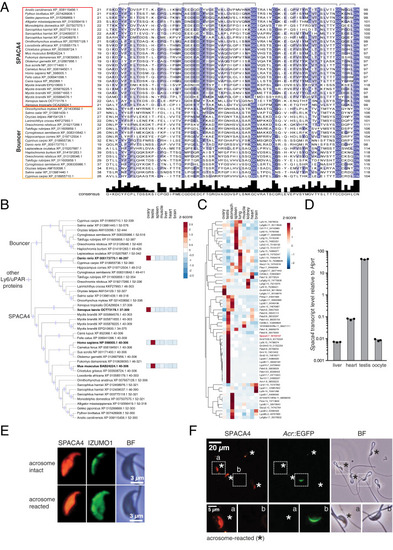
SPACA4 is expressed in murine sperm. (A) Protein sequence alignment of the mature domain of SPACA4/Bouncer protein family members. Shown is the mature three-finger domain of SPACA4 proteins (red box) and Bouncer proteins (orange box) lacking the N-terminal signal peptide and the carboxyl-terminal GPI-anchoring sequence. Amino acid divergence among different species is indicated based on the percent amino acid identity (blue shading); predicted cysteine bridges (gray) are shown above the alignment. Adapted from ref. 42. (B) Mammalian SPACA4 is the homolog for fish Bouncer and is expressed specifically in testis. Part of a maximum-likelihood phylogenetic tree based on Ly6/uPAR protein sequence alignments across vertebrates showing SPACA4 and Bouncer (refer to SI Appendix, Fig. S2A for the full tree; adapted from ref. 42). Branches supported by ultrafast bootstrap values (≥95%) are marked with a blue dot. Z-scores of averaged gene expression values across adult tissues are shown on the right for Danio rerio (11), Xenopus laevis (75), Mus musculus (73), and Homo sapiens (www.gtexportal.org). While Bouncer/Spaca4 mRNAs are expressed in oocytes in fish and frogs, mammalian Spaca4 mRNAs are expressed in testis. (C) Mouse Ly6/uPAR genes show diverse expression patterns across adult tissues. The heatmap is color coded based on z-scores of the normalized gene expression values (average of the square root) of RNA-Seq data from murine adult tissues (73). The clustering and dendrogram (Left) are based on expression scores. Spaca4 is highlighted in red. Numbers behind gene names indicate chromosome location in mice. (D) Mouse Spaca4 mRNA is expressed in the male germline. RT-qPCR from murine cDNA from different adult tissues reveals enrichment of Spaca4 specifically in mouse testis. Primers amplifying Hypoxanthineguanine phosphoribosyl transferase (Hprt) were used as a control. (E) Mouse SPACA4 protein localizes to the sperm head. Immunostaining of SPACA4 (red) and IZUMO1 (green) under permeabilizing conditions detects SPACA4 and IZUMO1 in the sperm head of wild-type B6D2F1 mice. In contrast to IZUMO1 that relocalizes after the acrosome reaction, SPACA4 does not change its localization. BF, brightfield image. (Scale bar, 3 µm.) (F) Murine SPACA4 localizes to the inner sperm membrane. Immunofluorescence staining of SPACA4 (red) in mouse spermatozoa under nonpermeabilizing conditions. Spermatozoa were derived from transgenic mice expressing EGFP under the control of the Acrosin promoter (65), labeling sperm with intact acrosomes (green). AR spermatozoa are highlighted by an asterisk (loss of green label). Boxed areas are shown below at higher magnification (a: AR sperm; b: spermatozoa with intact acrosome). SPACA4 is detected in AR spermatozoa (a) but not in spermatozoa with intact acrosomes (b). [Scale bar, 20 µm (Top) and 5 µm (Bottom).] |
|
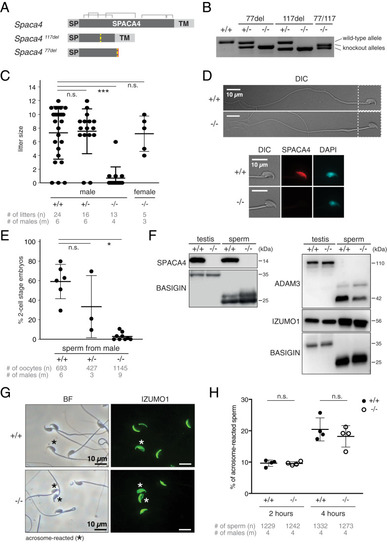
SPACA4 is required for efficient fertilization in male mice. ( |
|
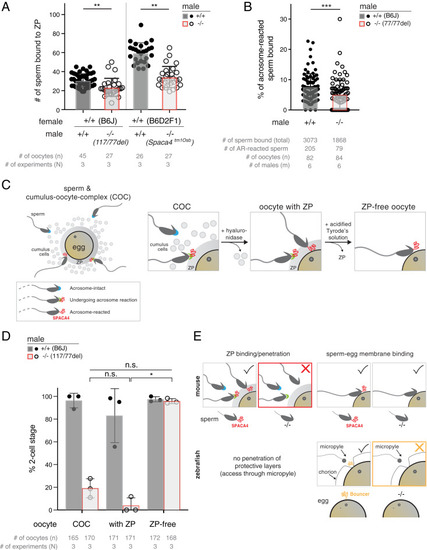
SPACA4 enables sperm to bind to and penetrate the ZP. ( |