- Title
-
Transcriptomics Profiling Identifies Cisplatin-Inducible Death Receptor 5 Antisense Long Non-coding RNA as a Modulator of Proliferation and Metastasis in HeLa Cells
- Authors
- Gurer, D.C., Erdogan, İ., Ahmadov, U., Basol, M., Sweef, O., Cakan-Akdogan, G., Akgül, B.
- Source
- Full text @ Front Cell Dev Biol
|
Cisplatin-induced apoptosis and proliferation in HeLa cells. |
|
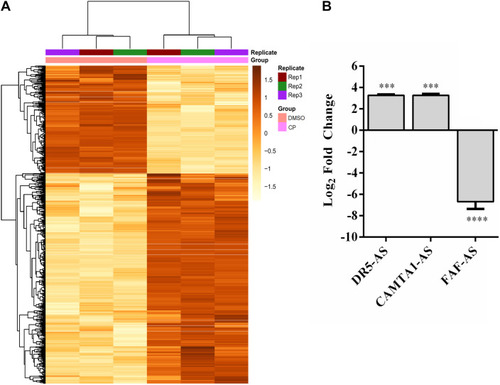
Differentially expressed lncRNAs in cisplatin-treated HeLa cells. |
|
Genomic location of DR5-AS and its expression in cisplatin-treated HeLa cells. |
|
DR5-AS knockdown modulates cell morphology in HeLa cells. |
|
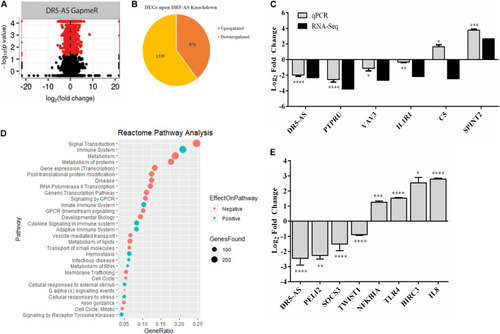
Figure 5. DR5-AS-knockdown-mediated perturbations in gene expression. Total RNAs isolated from HeLa cells transfected with negative GapmeR or DR5-AS GapmeR were subjected to RNA-seq. A volcano plot and pie chart of differentially expressed genes after knockdown are presented in (A,B), respectively. (C) qPCR validation of some genes differentially expressed in DR5-AS knockdown HeLa cells. (D) Reactome Pathway analysis of genes differentially expressed in HeLa cells transfected with DR5-AS GapmeR. (E) qPCR analyses of immune system-related genes as determined to be differentially expressed based on the Reactome pathway analysis. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ****p ≤ 0.0001. |
|
DR5-AS knockdown modulates proliferation and cell cycle in HeLa cells. |
|
Cisplatin- and TRAIL-induced cellular changes in DR5-AS knockdown HeLa cells. |
|
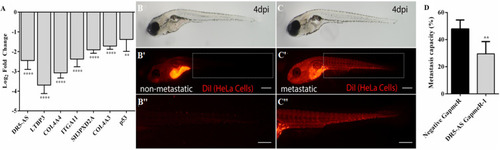
Analysis of metastasis rate in zebrafish xenograft assay. |