- Title
-
Identification and functional modelling of plausibly causative cis-regulatory variants in a highly-selected cohort with X-linked intellectual disability
- Authors
- Bengani, H., Grozeva, D., Moyon, L., Bhatia, S., Louros, S.R., Hope, J., Jackson, A., Prendergast, J.G., Owen, L.J., Naville, M., Rainger, J., Grimes, G., Halachev, M., Murphy, L.C., Spasic-Boskovic, O., van Heyningen, V., Kind, P., Abbott, C.M., Osterweil, E., Raymond, F.L., Roest Crollius, H., FitzPatrick, D.R.
- Source
- Full text @ PLoS One
|
(A) A diagrammatic summary of the experimental pipeline followed in this paper. (B) Schematic showing the genomic region of the six genomic variants in the five probands (S19, S24, S43, S3 and S31) indicating the location of the XLID-associated CRE variants along with their predicted target genes indicated in red, genomic coordinates from h19/GRCh37 genome build. The variants highlighted by grey box were used to make mouse models. |
|
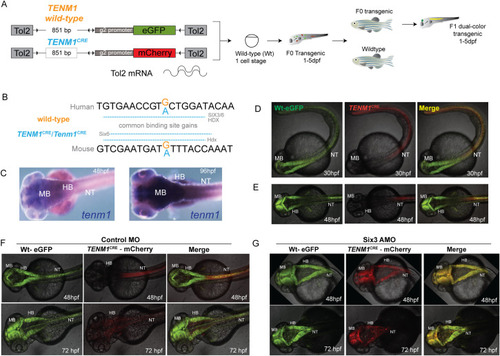
(A) A diagrammatic summary of the dual color fluorescence assay used in this study. The size of the human EXPRESSION / LABELING:
PHENOTYPE:
|
|
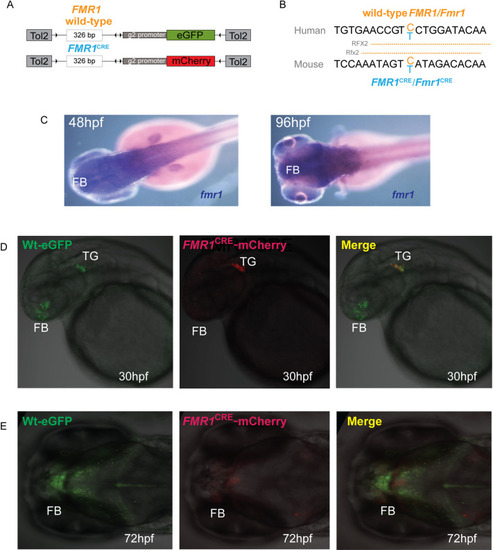
(A) A diagrammatic summary of the dual color fluorescence assay plasmid constructs used in this study. The size of the human PHENOTYPE:
|
|
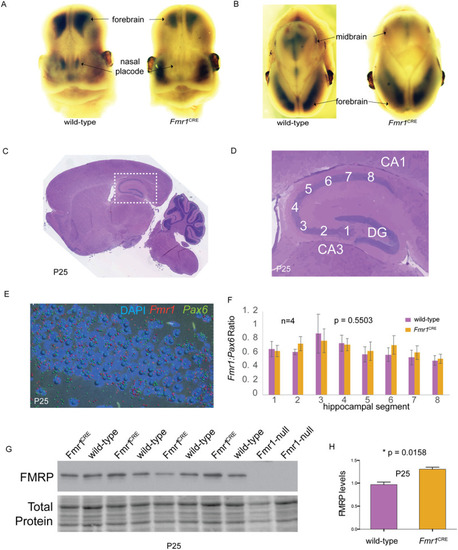
Frontal (A) and saggital (B) views of 13.5GD embryonic mouse heads following whole-mount |
|
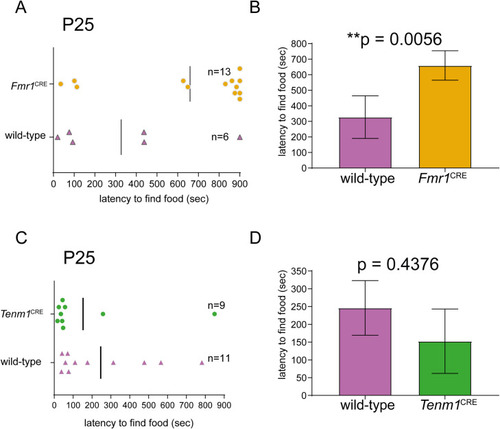
(A, B) The mice hemizygous for the variant in |
|
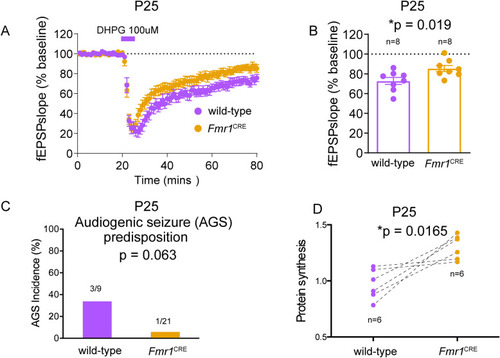
(A) Comparison of mGluR-dependent long-term depression (LTD) in CA3-CA1 components of the hippocampus of eight |
|
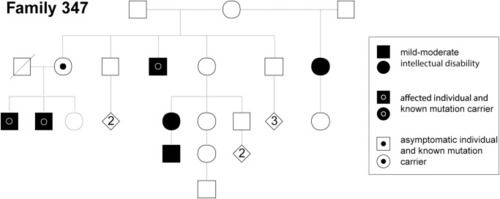
Pedigree of Family 347 of which individual S3 is a member showing segregation of the mutation affecting |