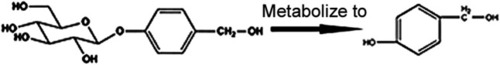- Title
-
P- Hydroxybenzyl Alcohol Alleviates Oxidative Stress in a Nonalcoholic Fatty Liver Disease Larval Zebrafish Model and a BRL-3A Hepatocyte Via the Nrf2 Pathway
- Authors
- An, J., Cheng, L., Yang, L., Song, N., Zhang, J., Ma, K., Ma, J.
- Source
- Full text @ Front Pharmacol
|
Chemical structure of GAS and HBA as well as metabolization of GAS into HBA |
|
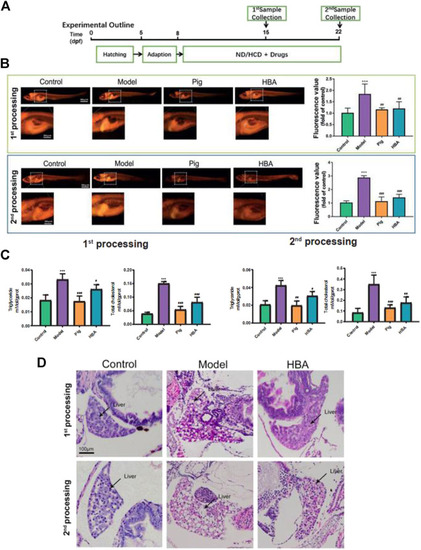
Effect of HBA in regulating lipid metabolism on an HCD-Induced Larval Zebrafish Model in two processes. |
|
Effect of HBA in oxidative stress on HCD-Induced Larval Zebrafish Model in two processes. |
|
Liver mRNA Expression Changes HBA on the Larval Zebrafish Model. |
|
The effect of HBA on FFA-induced BRL-3A cell |