- Title
-
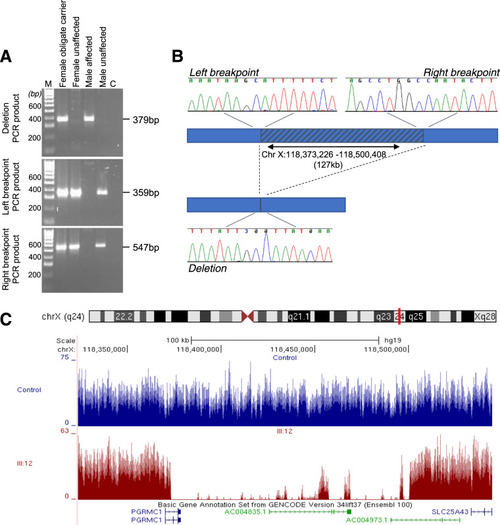
A 127 kb truncating deletion of PGRMC1 is a novel cause of X-linked isolated paediatric cataract
- Authors
- Jones, J.L., Corbett, M.A., Yeaman, E., Zhao, D., Gecz, J., Gasperini, R.J., Charlesworth, J.C., Mackey, D.A., Elder, J.E., Craig, J.E., Burdon, K.P.
- Source
- Full text @ Eur. J. Hum. Genet.
|
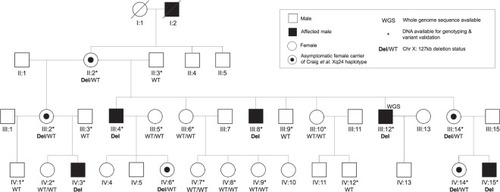
The previously studied CRVEEH66 family [ |
|
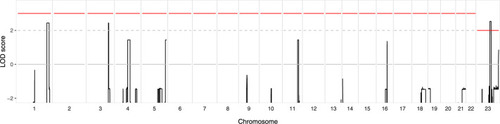
Multipoint LOD scores ( |
|
|
|
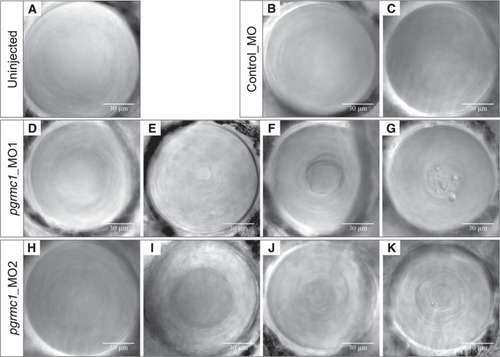
Representative DIC lens images from PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|