- Title
-
Klebsiella virus UPM2146 lyses multiple drug-resistant Klebsiella pneumoniae in vitro and in vivo
- Authors
- Assafiri, O., Song, A.A., Tan, G.H., Hanish, I., Hashim, A.M., Yusoff, K.
- Source
- Full text @ PLoS One
|
(a) Plaque morphology of UPM2146 was uniformly distributed after overnight incubation at 37°C. (b) The maximum size of plaques ranged from 0.7–1 cm with halo zone of 0.5 cm forming around it, at 36 hours incubation, 37°C. |
|
100 nm scaled bar is shown. |
|
One step growth curve of UPM2146 taking 20 min of latent period, 5 min of rise period, and exponential phase of 10 min. Total time required to lyse its host was 25 min that is characteristic of lytic phages. The experiment was done in triplicates. |
|
Bars with different letters indicate significant difference at p<0.05 (Tukey HSD post-hoc test). Error bars are standard deviations (n = 3). |
|
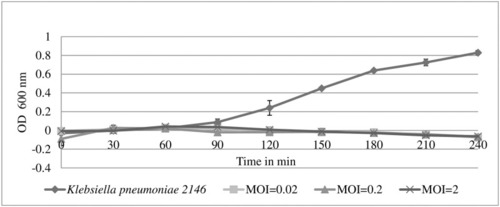
UPM2146 was able to lyse its host and clear the turbidity at 60 min for MOI of 0.02, 0.2 and 2. The bacterial count of control was 1.5±0.104 × 109 CFU/ml at an OD600nm of 0.8. The experiment was done in triplicates. |
|
This indicates the potential of UPM2416 in phage therapy. The circular genome map was constructed using SnapGene. |
|
Genomes of each phage is represented by linear visualization of their coding sequences (CDS) represented by arrows. The direction of the arrows indicates the direction of transcription of each CDS. The black vertical lines between the two genomic sequences indicate regions of shared similarities (89%) according to BLASTn. The intensity of these lines indicates the degree of similarity. This confirms that UPM2146 belongs to |
|
a. Phylogenic tree of UPM2146 show that the phylogenomic GBDP trees inferred using D6 formula and yielding average support of 58%, The numbers above branches are GBDP pseudo-bootstrap support values from 100 replications. The branch lengths of the resulting VICTOR tree are scaled in terms of D6 formula used. b. The evolution of UPM2146 of putative phage-encoded peptidoglycan binding protein had been drawn using phylogenic tree analysis using 1000 bootstrap, using the maximum likelihood (ML) method based on the Jones-Taylor-Thornton (JTT) model, the tree with the highest log likelihood (-1805.44) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 261 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [ |
|
(a.) zebrafish larvae were able to survive up to 24 h when infected with PHENOTYPE:
|
|
The phage UPM2146 managed to completely lyse |