- Title
-
Cardiac Regeneration and Tumor Growth-What Do They Have in Common?
- Authors
- Dicks, S., Jürgensen, L., Leuschner, F., Hassel, D., Andrieux, G., Boerries, M.
- Source
- Full text @ Front Genet
|
|
|
|
|
|
|
|
|
|
|
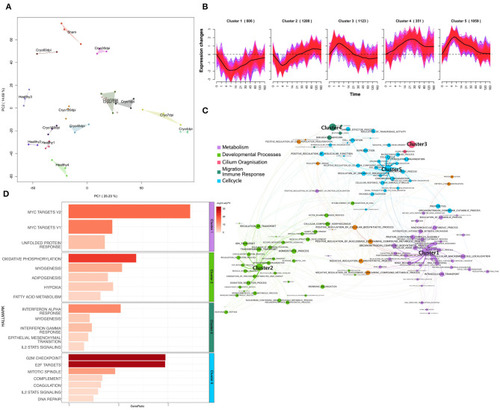
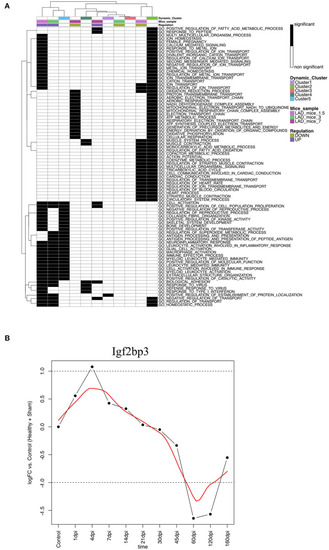
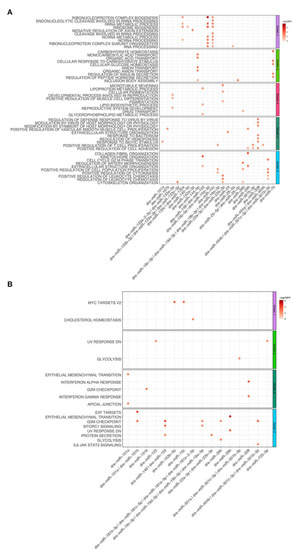
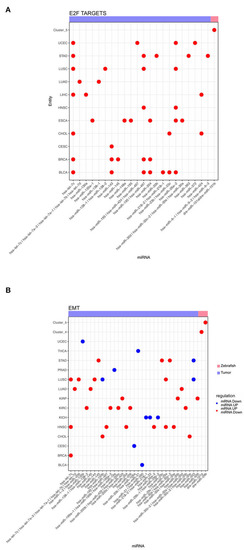
Dot plot of miRNA that significantly regulates the hallmarks |
|
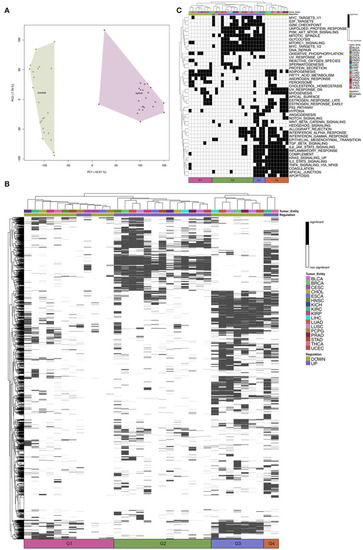
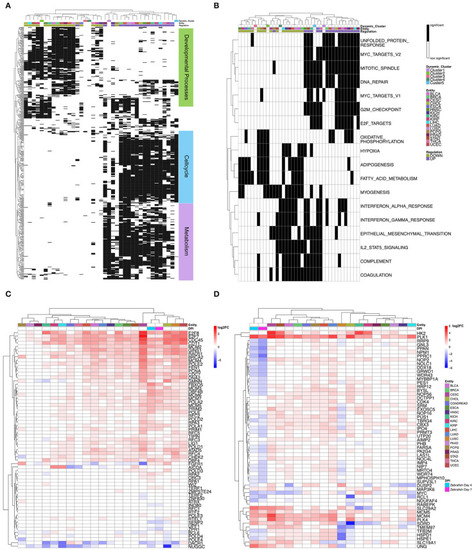
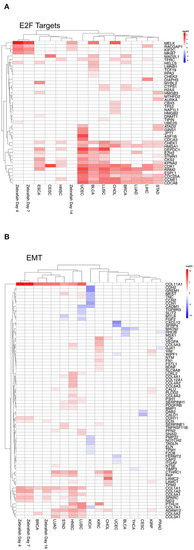
Heatmap of differential gene expression of significantly regulated mRNAs modulated by miRNA in the hallmarks |