- Title
-
In vivo biomolecular imaging of zebrafish embryos using confocal Raman spectroscopy
- Authors
- Høgset, H., Horgan, C.C., Armstrong, J.P.K., Bergholt, M.S., Torraca, V., Chen, Q., Keane, T.J., Bugeon, L., Dallman, M.J., Mostowy, S., Stevens, M.M.
- Source
- Full text @ Nat. Commun.
|
|
|
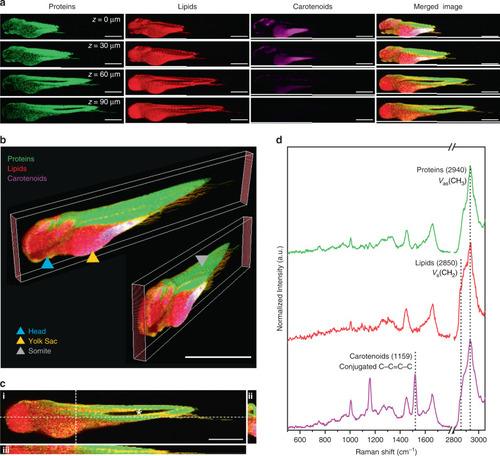
Confocal Raman spectroscopic imaging (cRSI) was used to image a fixed embryo ( |
|
Confocal Raman spectroscopic imaging (cRSI) was used to image tissue regions of interest at 3 days post fertilization, with a spatial resolution of 0.5–1 µm. Univariate analysis was performed by integrating over a wavenumber range corresponding to relevant biomolecules: collagen-rich regions at 918 ± 20 cm−1 (shown in yellow), DNA-rich regions at 789 ± 10 cm−1 (shown in blue), lipid-rich regions at 2850 ± 10 cm−1 (shown in red), cytochrome-rich regions at 1579 ± 15 cm−1 (shown in cyan). |
|
An established model was used in which zebrafish embryos were injected with |
|
|
|
Living zebrafish were imaged for the 12 h following a controlled stab wound performed at 3 days post fertilization. |