|
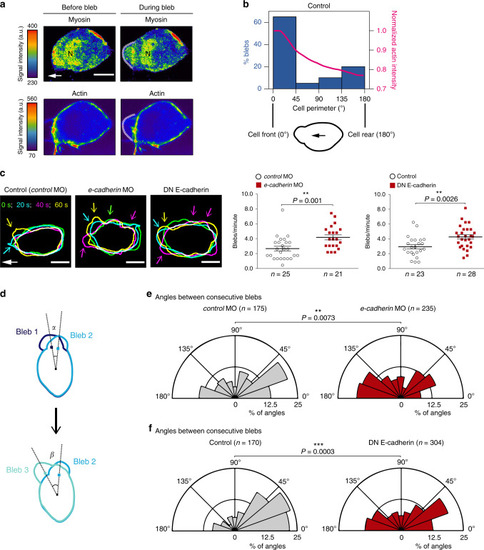
Myosin distribution in migrating PGCs and changes in the polarized generation of blebs upon E-cadherin knockdown.a Snapshots showing the distribution of myosin (Myl12.1-EGFP) and actin (LifeAct-mCherry) in a polarized PGC before and during bleb formation (see Supplementary Fig. 7a for an additional example). The snapshots are derived from Supplementary Movie 4, cell1, time points 0 s (before bleb) and 8 s (during bleb). White arrow indicates the direction of migration; N = myosin nuclear localization that might reflect a possible function of the protein in the nucleus (as described for human Myl12a also known as MRLC362); a.u. = arbitrary units; scale bars, 10 μm. The experiment was repeated three times. b Graph: percentage of blebs initiating at different angles around the cell perimeter (left y-axis, blue columns) and normalized actin intensity (right y-axis, magenta line) of Control cells (for cells expressing the DN E-cadherin and analyzed in a similar way see Supplementary Fig. 8c). The schematics shows the cell perimeter of a polarized PGC where 0° represents the cell front and 180° the cell rear. The black arrow indicates the direction of migration. A total number of 4 representative cells and 20 blebs were analyzed. c Left panels: overlays of the cell contours of three representative polarized PGCs at four time points, presenting bleb formation over 60 s. Time points are color-coded as indicated. Colored arrows point at blebs formed at time points of the corresponding colors. White arrow indicates the direction of migration; scale bars, 10 μm; s = seconds. The cell contours are derived from the movies in Supplementary Movie 6 (control MO: 0, 20, 40, 60 s; e-cadherin MO: 20, 40, 60, 80 s; DN E-cadherin: 15, 35, 55, 75 s). Graphs present the number of blebs per minute in polarized, motile PGCs. n = number of cells from 6 and 5 independent experiments for e-cadherin morpholino experiments and DN E-cadherin experiments, respectively; mean ± s.e.m.; P value: two-tailed Mann–Whitney U-test for morpholino and two-sided Student’s t-test for DN E-cadherin. d Schematics explaining the measurements provided in e and f. The angles between consecutive blebs were derived by measuring the angle between the two lines that connect the centre of the cell with the point of bleb initiation at a certain time point (for example, time point 1, bleb 1) and the site of initiation of the next bleb (for example, time point 2, bleb 2). e, f Polar plots show the distribution of angles between consecutive blebs in control cells (gray polar plots) and cells with manipulated E-cadherin function (red polar plots). n = number of angles obtained from 7 independent repeats for morpholino experiments (26 cells for control MO and 24 cells for e-cadherin MO) and 6 independent repeats for the DN E-cadherin experiments (23 cells for Control and 31 cells for DN E-cadherin). The time-lapse videos for characterizing blebbing were captured at 500 ms time intervals. P values: two-tailed Kolmogorov–Smirnov test. Source data are provided as a Source Data file.
|