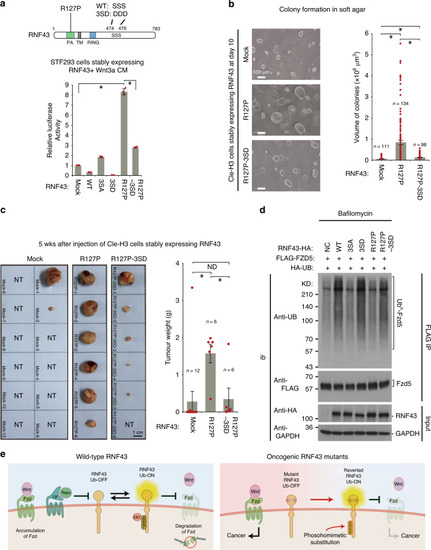
|
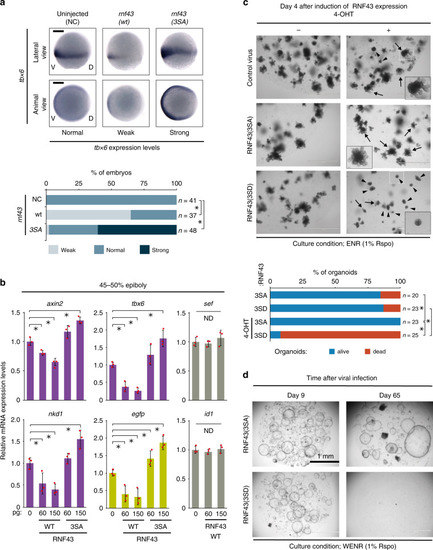
RNF43 phosphorylation regulates morphogenesis and ISC maintenance.a Expression of endogenous target genes of Wnt/β-catenin signalling was evaluated using in situ hybridisation following expression of RNF43 phospho-mutant forms in zebrafish embryos at 8 h post-fertilisation (hpf) (75% epiboly). Scale bars, 200 μm. Asterisks indicate significant differences (P < 0.05, one-way ANOVA, n = 37–48 as indicated) from NC embryos. b Expression change of Wnt/β-catenin, FGF and BMP signalling target genes with RNF43 was evaluated using qPCR at 5.3 hpf (45–50% epiboly). Endogenous Wnt target genes, artificial Wnt reporter gene and non-Wnt target genes are shown in purple, green and grey, respectively. Expression of each gene in uninjected embryos was set to 1 (mean ± sd). Red circles indicate individual values of each sample. Asterisks indicate significant differences (P < 0.05, one-way ANOVA, n = 3, biological replicates with pools of 25–30 embryos) from uninjected embryos. ND indicates no significant difference. c Short-term development of intestinal organoids was examined at 4 days following induction of RNF43 phospho-mutant forms. White dashed boxes denote the area enlarged in black boxes. Arrows, healthy organoids. Arrowheads, dead organoids. Asterisks indicate significant differences (P < 0.05, one-way ANOVA, n = 20–25 as indicated) between groups. d Long-term ISC maintenance was evaluated after 65 days with two passages in organoids expressing RNF43 phospho-mutant forms. Scale bars in (c, d), 1 mm.
|