- Title
-
Effect of Ataluren on dystrophin mutations
- Authors
- Berger, J., Li, M., Berger, S., Meilak, M., Rientjes, J., Currie, P.D.
- Source
- Full text @ J. Cell. Mol. Med.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
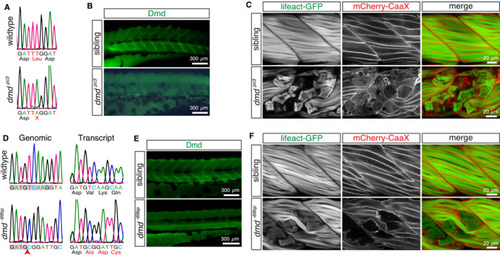
Novel dystrophin‐deficient mutants. A, A > T substitution within exon 34 of EXPRESSION / LABELING:
PHENOTYPE:
|
|
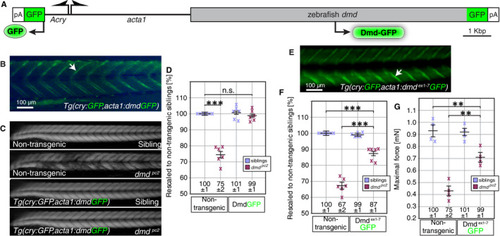
The dystrophin isoform Dmd∆ex1‐7 lacking exons 1‐7 is partially functional. A, Schematic of the construct used to generate the transgenic line |