- Title
-
Generating Stable Knockout Zebrafish Lines by Deleting Large Chromosomal Fragments Using Multiple gRNAs
- Authors
- Kim, B.H., Zhang, G.
- Source
- Full text @ G3 (Bethesda)
|
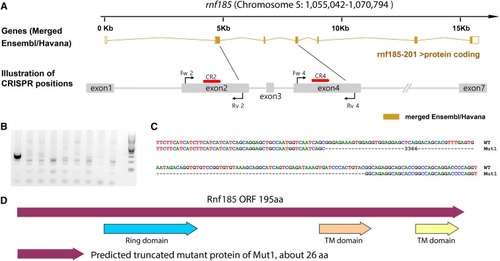
Multiple gRNAs targeting |
|
LKO mutations induced by multiple gRNAs targeting the |
|
LKO mutations induced by multiple gRNAs targeting the |