- Title
-
Parallel Reaction Monitoring reveals structure-specific ceramide alterations in the zebrafish
- Authors
- Zhang, T., Trauger, S.A., Vidoudez, C., Doane, K.P., Pluimer, B.R., Peterson, R.T.
- Source
- Full text @ Sci. Rep.
|
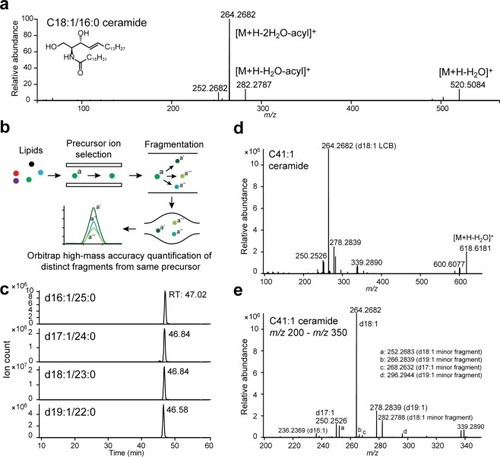
A Parallel Reaction Monitoring-based approach for ceramide quantification. ( |
|
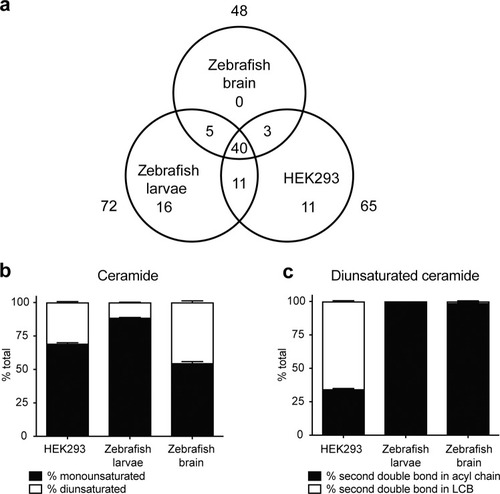
Ceramide composition of zebrafish brain, larvae and HEK239 cells. ( |
|
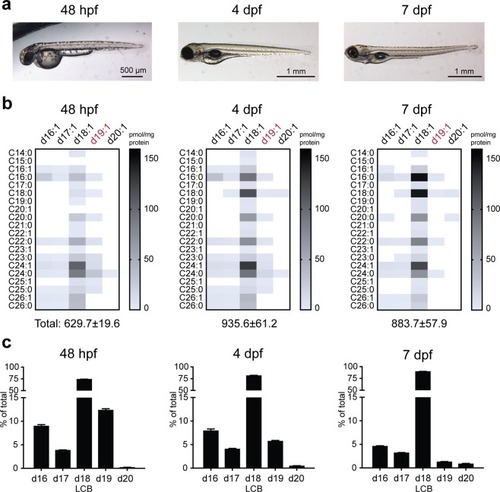
Ceramide regulation during zebrafish embryogenesis. ( |
|
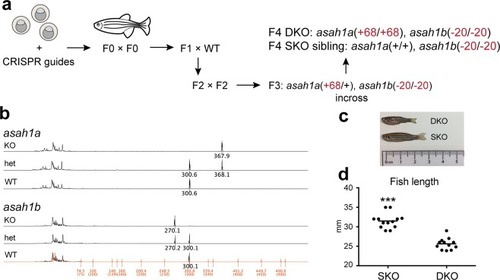
A zebrafish model of Farber disease. ( PHENOTYPE:
|
|
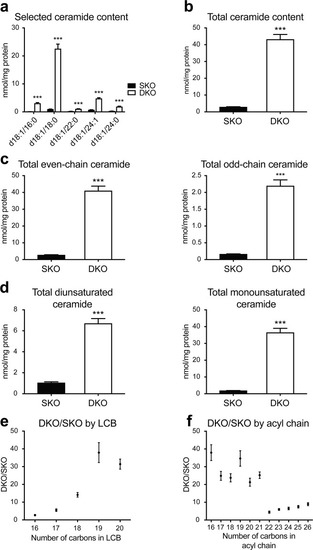
Altered ceramide distribution in Farber disease zebrafish. ( PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

Unillustrated author statements PHENOTYPE:
|