- Title
-
Zebrafish Vestigial Like Family Member 4b Is Required for Valvulogenesis Through Sequestration of Transcription Factor Myocyte Enhancer Factor 2c
- Authors
- Xue, C., Liu, X., Wen, B., Yang, R., Gao, S., Tao, J., Zhou, J.
- Source
- Full text @ Front Cell Dev Biol
|
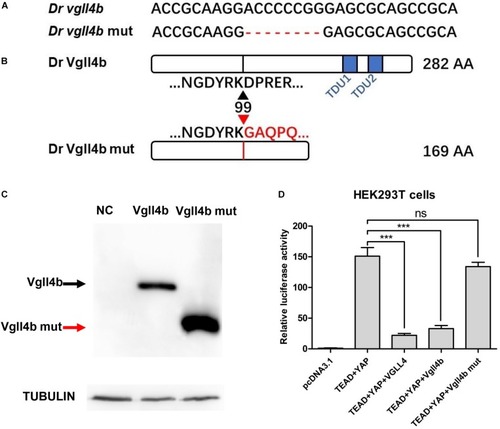
The establishment of a zebrafish |
|
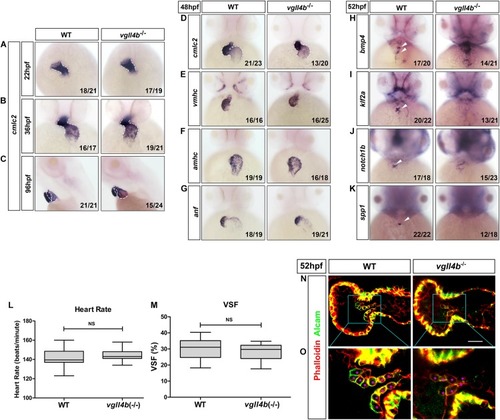
Deficiency of zebrafish |
|
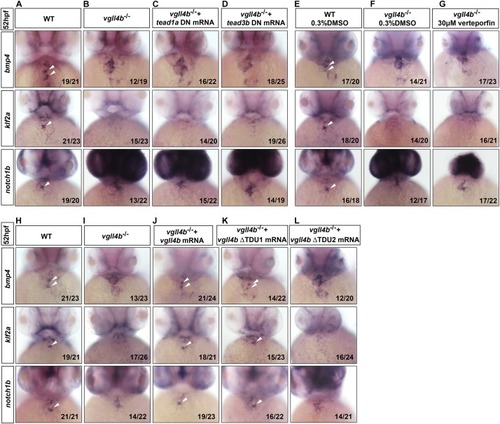
Tead is not involved in the impaired valvulogenesis in |
|
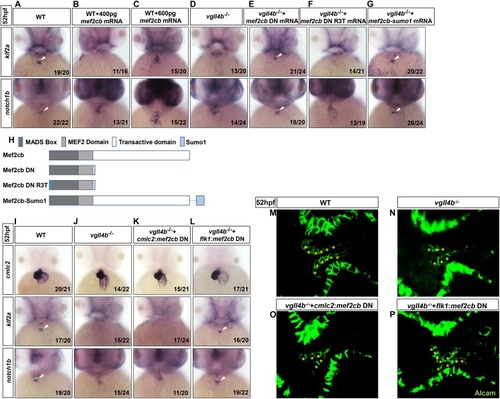
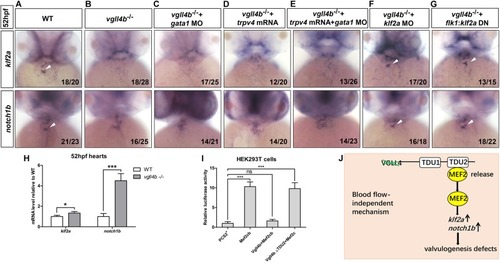
Aberrant activation of Mef2c due to the disruption of Vgll4b-Mef2c complex, accounts for the valvulogenesis defects in vgll4b mutants. |
|
The failure of |