- Title
-
Regeneration Rosetta: An Interactive Web Application To Explore Regeneration-Associated Gene Expression and Chromatin Accessibility
- Authors
- Rau, A., Dhara, S.P., Udvadia, A.J., Auer, P.L.
- Source
- Full text @ G3 (Bethesda)
|
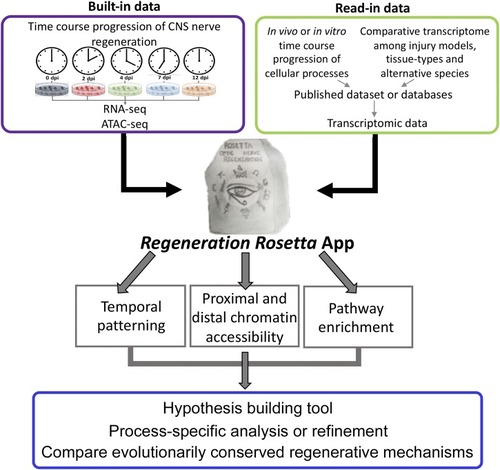
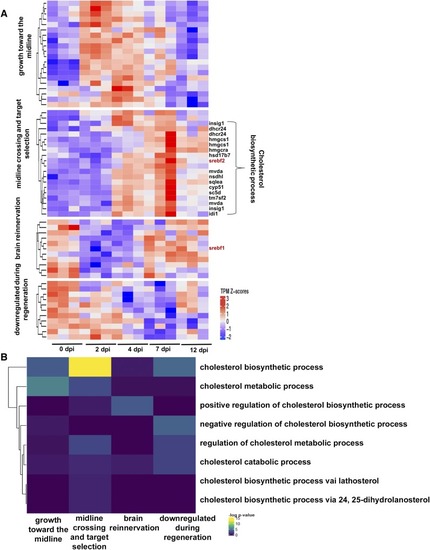
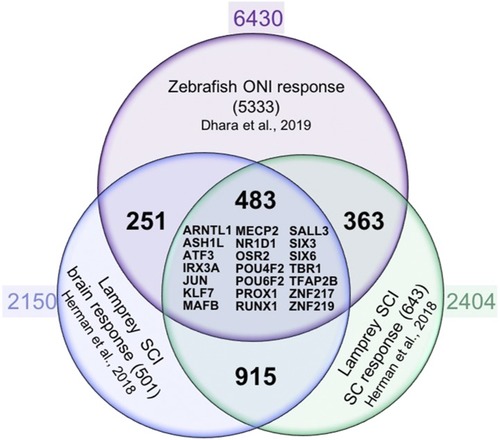
Workflow of |
|
|
|
|